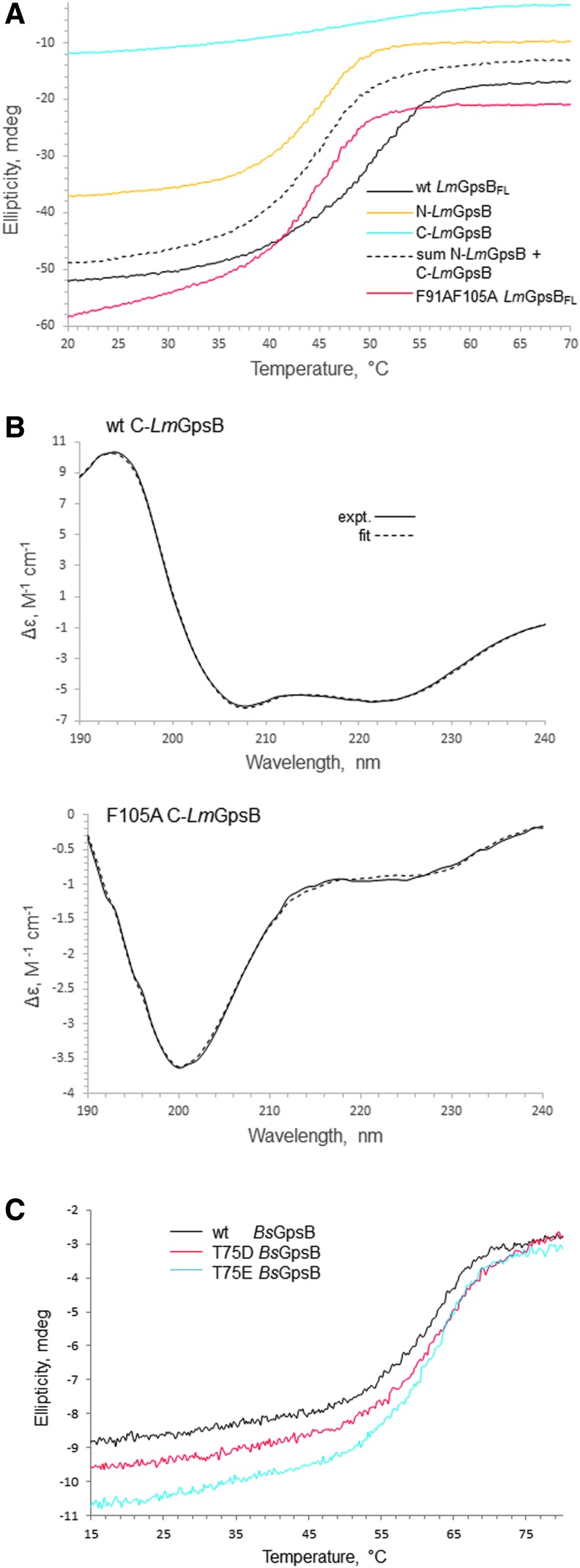
FIG. 6.
Circular dichroism spectra. (A) Thermal denaturation of LmGpsB (black line), N-LmGpsB (orange line), C-LmGpsB (cyan line), and LmGpsBF91AF105A (red line), monitored by circular dichroism; the dashed line represents the summation of the individual thermal melts measured for N-LmGpsB and C-LmGpsB. Unfolding of the secondary structure is observed by monitoring ellipticity at 222 nm. The F91AF105A mutation reduces the stability of LmGpsB. (B) Circular Dichroism spectra of wild-type C-LmGpsB (top) and C-LmGpsBF105A (bottom panel). The dashed spectrum represents the reconstructed spectra after fitting the secondary structural content with the program CDSStr (30). (C) Thermal denaturation of wt BsGpsB (black line), BsGpsBT75D (red line), and BsGpsBT75E (cyan line) monitored by circular dichroism ellipticity at 222 nm. The midpoint of the unfolding transition is similar for the three proteins (c. 62°C).