Fig. 1.
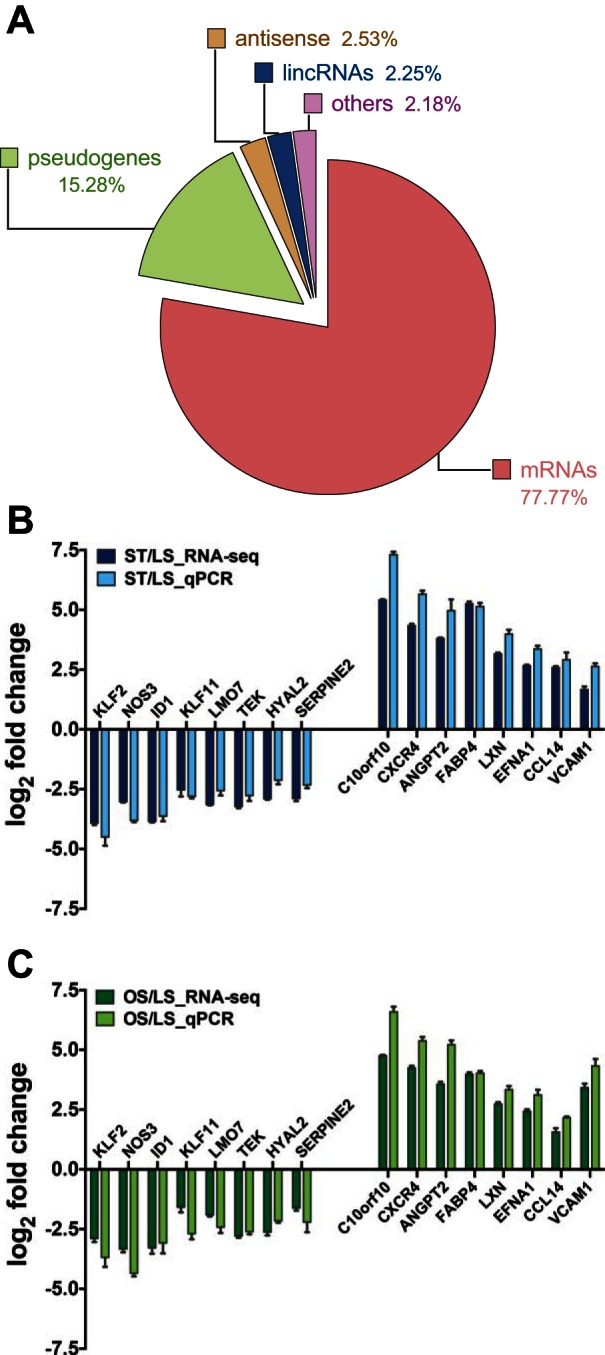
Summary of RNA-seq detected transcripts and quantitative real-time PCR (qPCR) confirmation. A: a pie chart showing the distribution of all detected genes as messenger RNAs (mRNAs), pseudogenes, antisense, long intervening noncoding RNAs (lincRNAs), and others. qPCR confirmation of representative genes detected by RNA-seq under static conditions (ST, B) and oscillatory sheer stress (OS, C) including Krüppel-like factor 2 (KLF2), nitric oxide synthase 3 (NOS3), inhibitor of DNA binding 1 (ID1), KLF11, LIM domain 7 (LMO7), TEK tyrosine kinase (TEK), hyaluronoglucosaminidase 2 (HYAL2), serpin peptidase inhibitor member 2 (SERPINE2), chromosome 10 open reading frame 10 (C10orf10), C-X-C chemokine receptor type 4 (CXCR4), angiopoietin-2 (ANGPT2), fatty acid binding protein 4 (FABP4), latexin (LXN), Ephrin-A1 (EFNA1), chemokine (C-C motif) ligand 14 (CCL14), vascular cell adhesion molecule 1 (VCAM1) (n = 4, respectively).
