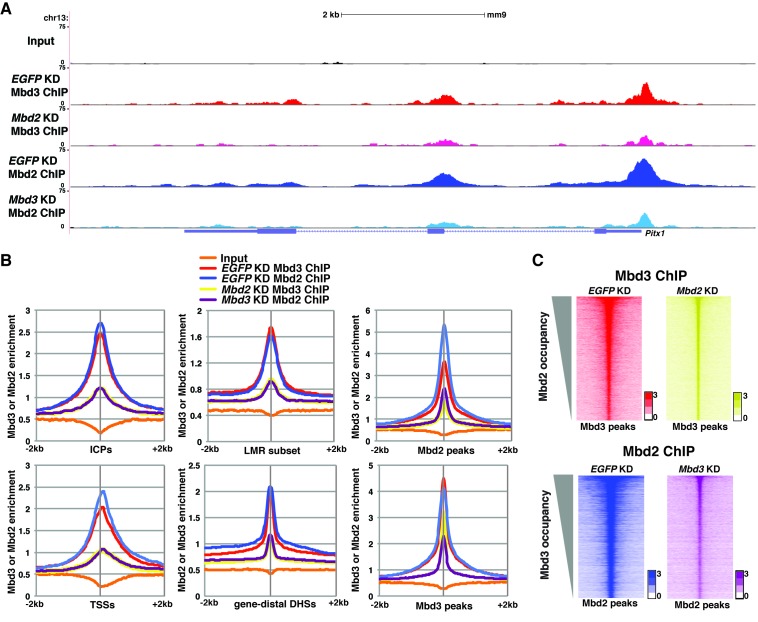
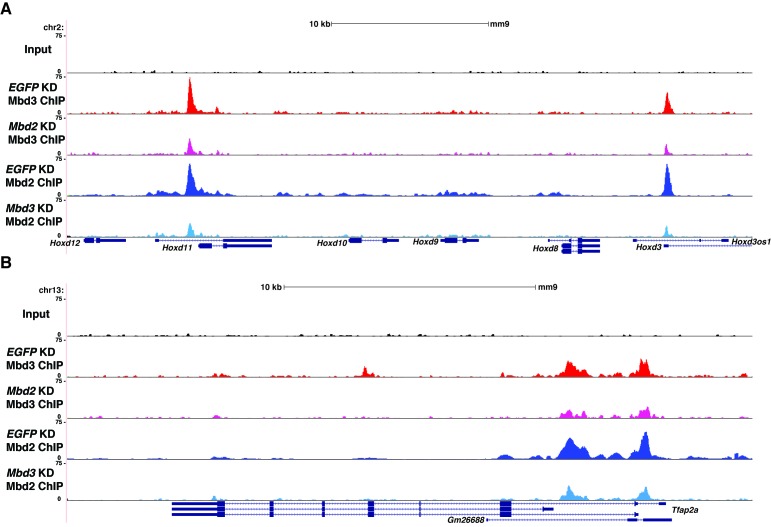
Figure 5. Mbd3 and Mbd2 are required for each other’s binding in ES cells.
(A) Genome browser tracks of ChIP-seq experiments examining Mbd3 or Mbd2 occupancy in control (EGFP KD), Mbd2 KD, or Mbd3 KD ES cells over one example locus (Pitx1). (B) Aggregation plots of Mbd3 or Mbd2 ChIP-seq data showing occupancy over ICPs (top left panel), annotated TSSs (bottom left panel), LMR subset (top middle panel), gene-distal DHSs (bottom middle panel), Mbd2 peaks (top right panel), and Mbd3 peaks (bottom right panel) ± 2 kb in control (EGFP KD), Mbd3 KD, or Mbd2 KD ES cells. (C) Heatmaps of Mbd3 enrichment over Mbd3 binding sites ± 2 kb sorted by Mbd2 occupancy (top panel) and Mbd2 enrichment over Mbd2 binding sites ± 2 kb sorted by Mbd3 occupancy (bottom panel) in control (EGFP KD), Mbd2 KD, or Mbd3 KD ES cells. The profiles shown in the browser tracks, aggregation plots, and heatmaps represent the average of two biological replicates.