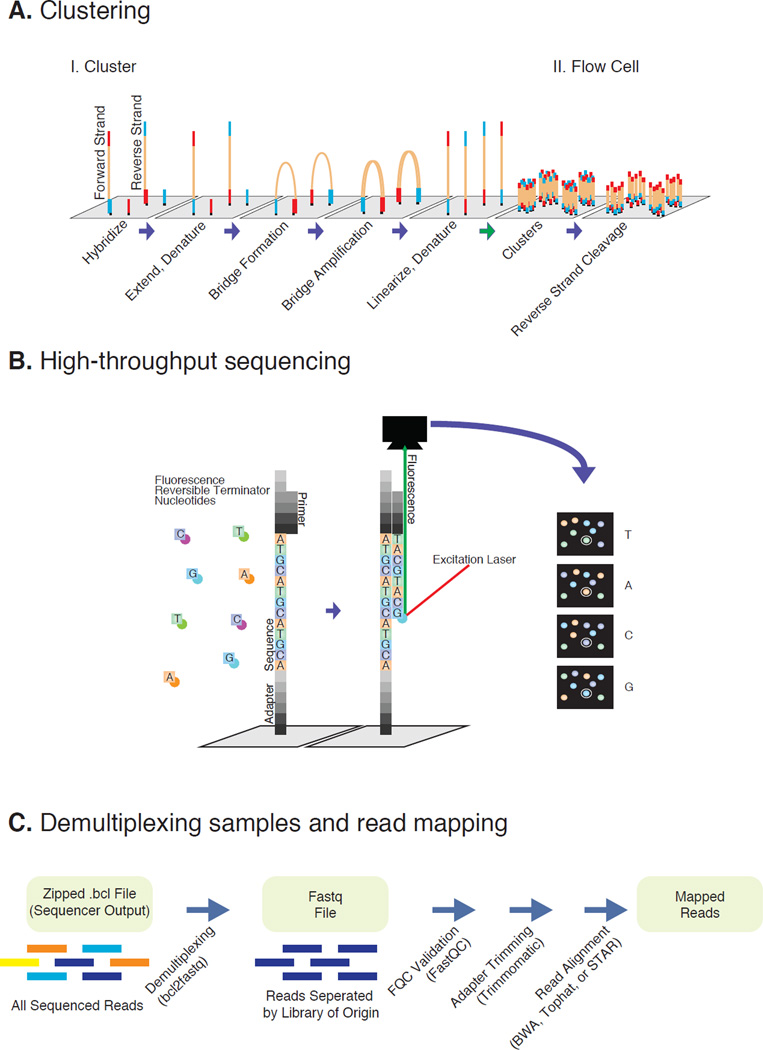
Figure 2. Illumina sequencing and data processing workflow.
A. Denaturated NGS library fragments are flowed across a flow cell and hybridize on a lawn of complementary Illumina adapter oligos. Complementary fragments are extended, amplified via bridge amplification PCR, and denaturated, resulting in clusters of identical single-stranded library fragments. B. Fragments are primed and sequenced utilizing reversible terminator nucleotides. Base pairs are identified after laser excitation and fluorescence detection. C. Raw data is demultiplexed into individual libraries and assessed for quality. Removing adapter reads reduces technical noise. Finally reads are aligned onto assembly of interest.