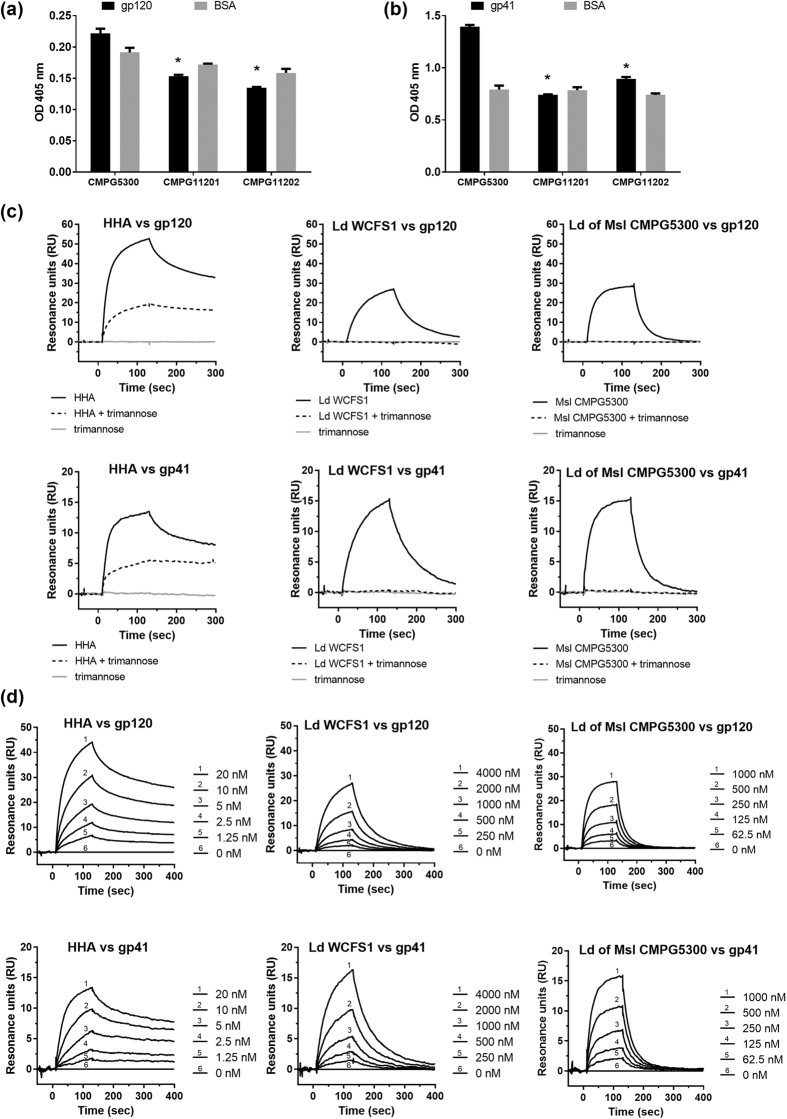
Figure 5. HIV-1 gp120 and gp41 binding of bacterial cells and the purified lectin domain of Cmpg5300.05_29.
(a) Binding of biotin-labeled cells of the wild-type strain L. plantarum CMPG5300, mutant CMPG11201 and complemented strain CMPG11202 to HIV-1 gp120. The OD at 405 nm reflects the binding efficiency of the bacterial strains. BSA was used as a negative control. (b) Binding of biotin-labeled cells of the wild-type strain L. plantarum CMPG5300, mutant CMPG11201 and complemented strain CMPG11202 to HIV-1 gp41. The OD at 405 nm reflects the binding efficiency of the bacterial strains. BSA was used as a negative control. (c) Binding of the lectin domain of CMPG5300 (Ld CMPG5300) to HIV-1 gp120 (solid lines) and HIV-1 gp41 (solid lines) in terms of Resonance units as analyzed via SPR. The purified lectin domain of Msa of L. plantarum WCFS1 (Ld WCFS1) was used as control (solid lines) and HHA was used as positive control (solid lines). Upon addition of competitor trimannose Manα(1-3)Manα(1-6)Man to the lectin domains, binding to HIV-1 gp120 was abolished (dotted lines). The concentrations of the lectins used are 20 nM, 4 μM and 1 μM for HHA (left), lectin domain of Msa of L. plantarum WCFS1 (centre) and lectin domain of CMPG5300.05_29 of CMPG5300 (right), respectively. A concentration of 1 mM was used for trimannose. (d) Kinetic analysis for the interaction of the lectin domain of CMPG5300.05_29 with HIV-1 gp120 and gp41 using the lectin domain of Msa and HHA as control. Serial two-fold analyte dilutions (covering a concentration range from 0 to 20 nM for HHA, 0 to 4000 nm for Ld WCFS1 and from 0 to 1000 nM for Ld CMPG5300) were injected over the surface of the immobilized gp120 and gp41. A 1:1 binding model was applied to determine the kinetic parameters. The biosensor chip density was 115 RUs for gp120 and 45 RUs for gp41. The dataset comparisons (mutant pairwise to wild-type) are considered significant (p < 0.05 indicated with one asterisks). The dataset comparison (complemented mutant pairwise to the mutant strain) show no significant differences.