Fig. 2.
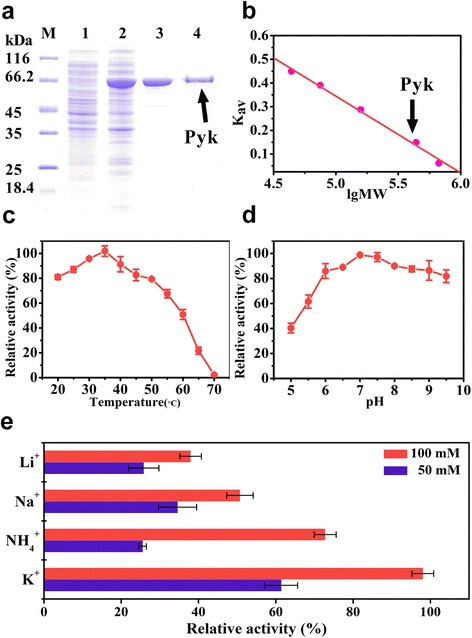
Enzymatic properties of Pyk2 from C. glutamicum. a SDS-PAGE of purified Pyk2. Lane 1, supernatant of crude extract from E. coli BL21(DE3)/pET-28a; Lane 2, supernatant of crude extract from E. coli BL21(DE3)/pET-28a-pyk2; Lane 3, Pyk2 purified by His-tagged affinity chromatography; and Lane 4, Pyk2 purified by ion exchange chromatography. b Molecular weight determination by gel filtration chromatography. The molecular weight of a protein could be determined from the calibration curve (plot of Kav versus the logarithm of molecular weight). The Kav value was calculated from the measured elution volume using the following equation: Kav = (Ve − Vo)/(Vc − Vo), where Ve is the elution volume, Vo is the column void volume, and Vc is the geometric column volume. The logarithm of the molecular weight was represented as lgMW. The standard proteins were shown as follows (●): 1, ovalbumin (44 kDa); 2, conalbumin (75 kDa); 3, aldolase (158 kDa); 4, ferritin (440 kDa); and 5, thyroglobulin (669 kDa). The native molecular weight of Pyk2 was determined to be a homohexamer of 430,881 (arrow). c The enzyme activity of Pyk2 at different temperatures. The maximum value of 2.19 ± 0.09 U/mg was set as relative 100%. d Enzyme activities of Pyk2 at various pHs (The maximum value of 2.43 ± 0.02 U/mg was set as relative 100%). e Enzyme activities of Pyk2 in the presence of various monovalent ions (The maximum value of 1.77 ± 0.05 U/mg was set as relative 100%). The enzyme was incubated in 100 mM Tris-HCl buffer (pH 7.4) and 5 mM Mn2+ with 50 mM or 100 mM monovalent ions. The data were derived from at least three replicate experiments, and the error bars represent the standard deviations
