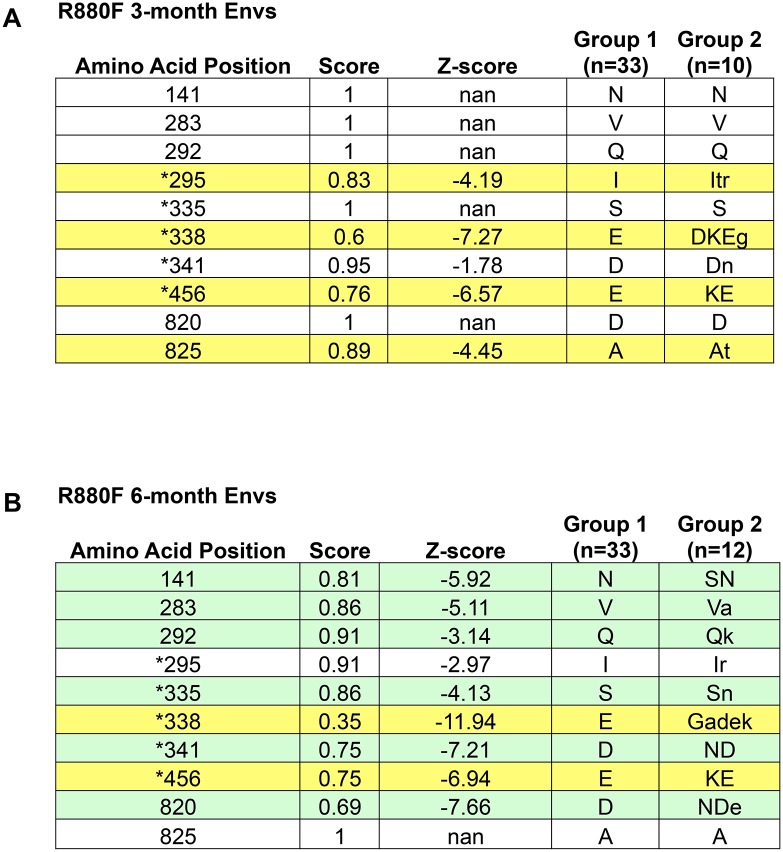
Fig 4. Validation of the Immunotype Diversity Index approach to quantify Env diversification using R880F.
Subject R880F was chosen to validate the Sequence Harmony approach because the earliest neutralizing antibody response was previously mapped to an epitope located at the base of the V3 loop [11]. Positions indicated with an asterisk were identified and evaluated in the previous study. Thirty-three SGA derived T/F Env amino acid sequences from R880F (Group 1) were aligned with 10 sequences from 3-months post-infection (A), or 12 sequences from 6-months post-infection (B) (Group 2). In (A), Sequence Harmony analysis identified four positions, highlighted yellow, that were significantly different (Z<-3) between the two populations at 3-months. Two of these positions were determined to be autologous neutralizing antibody escape mutations at 3-months via mutagenesis (295 and 338), whereas 456 could not be directly attributed to neutralizing antibody or cytotoxic T lymphocyte selective pressure. In (B), Sequence Harmony analysis identified two positions that remained significant from 3 months (highlighted yellow), six positions that became significant at 6 months (highlighted green), and two positions that lost significance at 6 months (not highlighted). Positions 335, 338, and 341 were all determined to be neutralizing antibody escape mutations at 6 months.