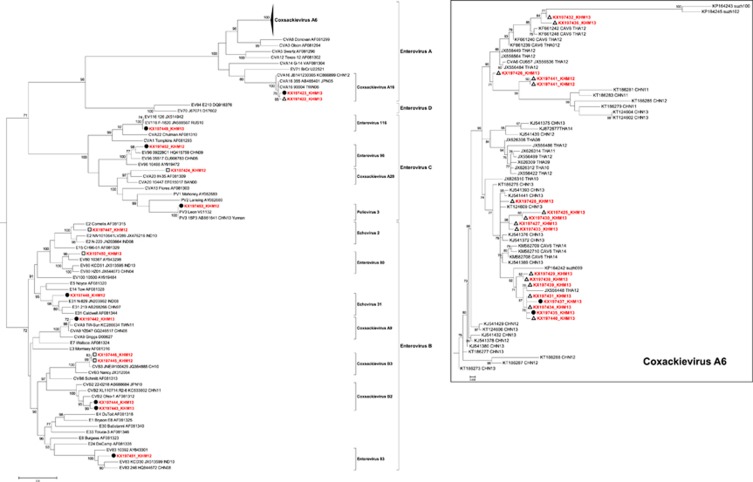
Figure 2.
Maximum likelihood (ML) phylogenetic tree of 140 VP1 sequences from all four enterovirus species (A, B, C and D). The tree was built using the ML method based on the GTR+G4 model. The robustness of nodes was assessed with 1000 bootstrap replicates. Bootstrap values <70 are not shown. The sequences isolated from Cambodian patients with different disease categories of severity are marked by white triangles for hand, foot and mouth disease, white rectangles for central nervous system involvement and black circles for cardiopulmonary failure.