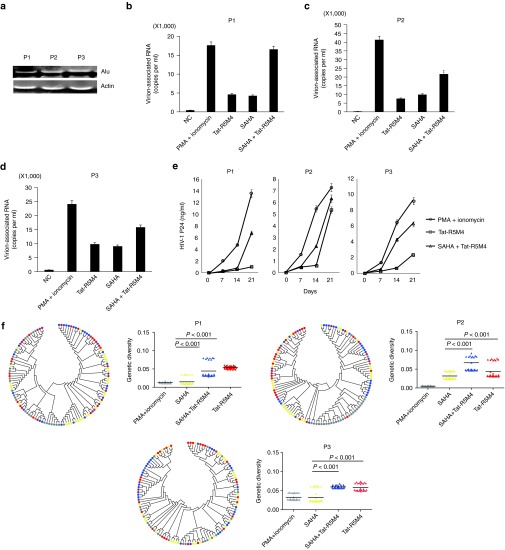
Figure 4.
Activation of the latently-infected CD4+ T-lymphocytes from HIV-1-infected individuals receiving suppressive cART by Tat-R5M4. (a) Proviruses in CD4+ T-lymphocytes isolated from HIV-1-infected individuals were detected by Alu-PCR. (b–d) The CD4+ T-lymphocytes isolated from HIV-1-infected individuals were cultured in the RPMI1640 conditioned medium and activated with phorbol myristate acetate (PMA) plus ionomycin, suberoylanilide hydroxamic acid (SAHA), Tat-R5M4, or SAHA plus Tat-R5M4. After 48 hours, the viral particles in supernatant were harvested and viral RNA was extracted and quantitatively analyzed by real-time reverse transcription-PCR. (e) The CD4+ T-lymphocytes were exposed to 200Gy X-ray irradiation and then co-cultured with the freshly-activated CD4+ T-lymphocytes from HIV-1-negative donors. The viral replication was monitored with the continuous detection of HIV-1 p24 antigen in the supernatant. (f) Genetic diversity analysis. HIV-1 strains were sampled from three virally activated subjects treated by PMA plus ionomycin (gray), SAHA (yellow), SAHA plus Tat (blue) and Tat (red) respectively, and 30 clones were sequenced in each case. Right panel: each triangle represents the genetic distance between one given clone and the relevant entire population, and the horizontal bar indicates the mean. Left panel: the bootstrap consensus trees were constructed based on HIV-1 sequences obtained from the corresponding patients annotated in the right panel. The two-tailed Mann-Whitney U-test was used to compare the genetic diversities between different groups.