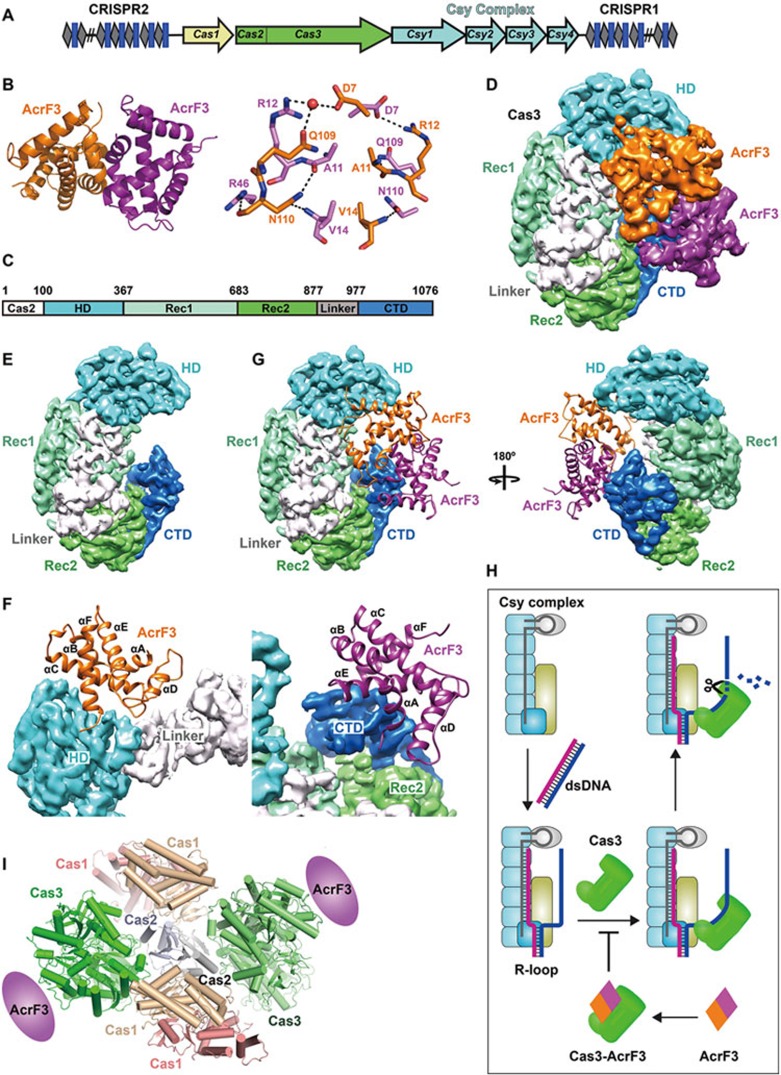
Figure 1.
Viral AcrF3 dimer binds to PaeCas3 protein, inactivating the bacterial CRISPR system. (A) Graphic representation of the P. aeruginosa CRISPR/Cas locus. The CRISPR locus consists of series of repeats (gray diamonds) separated by spacer sequences (blue rectangles). Four Csy genes are shown in cyan, Cas1 in yellow and the fused Cas2-Cas3 in green. (B) Overall crystal structure of AcrF3 (left panel) and the interactions between two monomers (right panel). (C) Domain organization of PaeCas3. (D) Architecture of the PaeCas3-AcrF3 complex. Cryo-EM density map of PaeCas3-AcrF3 complex at 4.2 Å resolution was segmented and colored according to subunit and domains. (E) Segmented PaeCas3 with domains color-coded. (F) Close-up view of interactions between PaeCas3 and AcrF3 dimer. One AcrF3 monomer interacts with HD domain and Linker of Cas3 (left panel); and the other AcrF3 monomer interacts with CTD domain of Cas3 (right panel). (G) AcrF3 dimer blocks the replaced non-complementary DNA access of Cas3. AcrF3 is shown as cartoon. (H) Model of AcrF3 inactivating CRISPR/Cas immune system. (I) Model of Cas1-Cas2-Cas3 complex.