Figure 4.
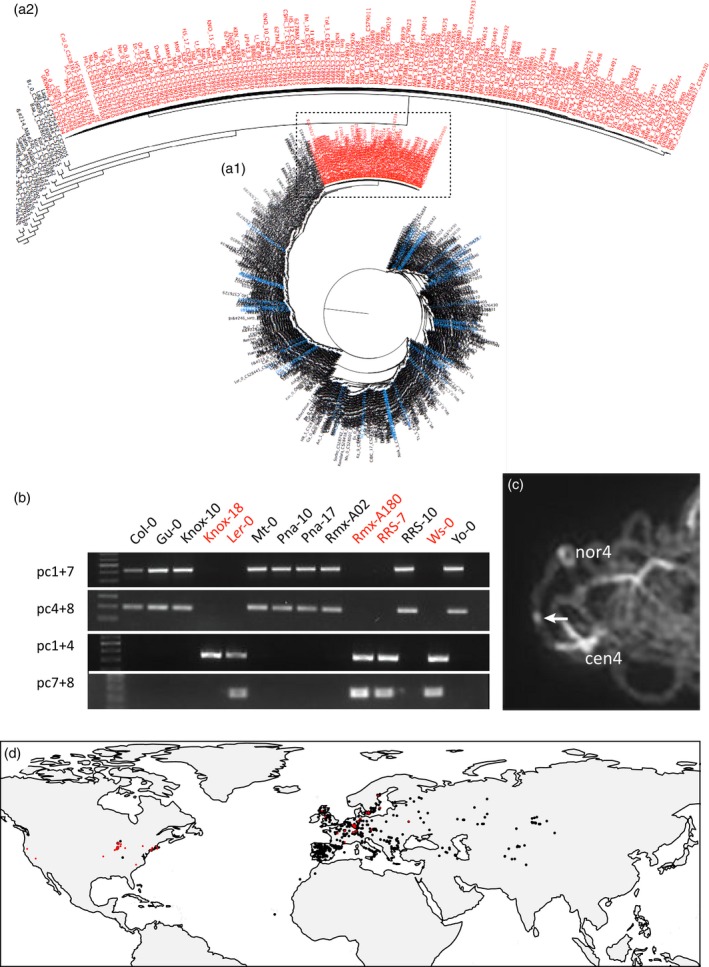
Identification of inversion accessions.
(a) Phylogenetic tree of all accessions from the 1001 genome database based on the 1.17 Mb inversion sequence. The inversion accessions (red names) form a monophyletic clade separated from all others. (a1) shows 1130 accessions, (a2) is a magnification of the clade with 132 inversion accessions.
(b) Identification of inversion accessions by PCR analysis of the inversion breakpoints using primers that flank the distal (pc1+7) and proximal (pc4+8) side of the inversion. The absence of the inversion breakpoints (accession name in red) gives rise to PCR fragments in case the primers pc1+4 or pc7+8 are used. The absence of a pc7+8 band in Knox‐18 is probably due to an aberrant nucleotide in this non‐inversion accession.
(c) DAPI staining of pachytene bivalent of accession Gu‐0 showing the heterochromatin knob hk4S (arrow) on chromosome 4 indicating the presence of the inversion.
(d) Global distribution of inversion (red) and non‐inversion accessions (black) from the 1001 Genomes Project. The inversion haplotype of 110 American inversion accessions is similar to UKSWO6‐333 (1), DIR‐9 (2) and NOZ‐6 (3) located in the UK and France.
