Figure 4.
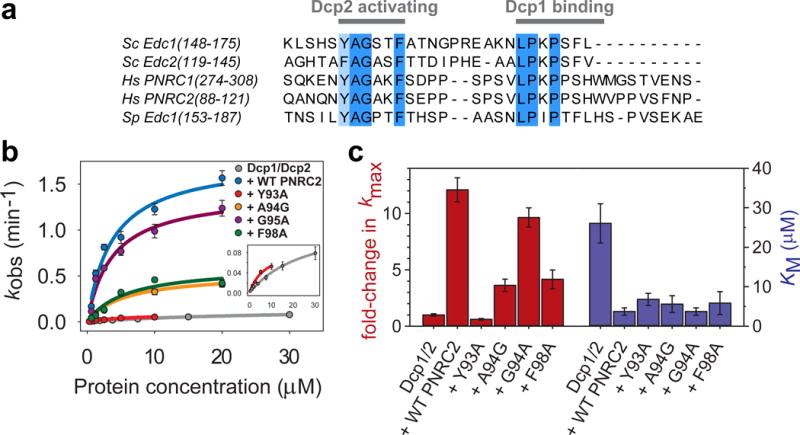
A conserved short-linear motif enhances the catalytic step of decapping. (a) Sequence alignment of decapping coactivators that share the tandem Dcp2-activating motif and Dcp1-binding motif. (b) Fits of kobs versus protein concentration under single-turnover conditions to determine kmax and KM for Dcp1–Dcp2 and WT or mutant complexes of Hs PNRC2(1-121) with Sp Dcp1–Dcp2(1-243). The single point mutation Y93A in the PNRC2 Dcp2-activating motif completely abolishes its ability to activate decapping chemistry. Errors are the standard error of individual fits to determine kobs. (c) Bar graph showing effects of PNRC2 and point mutants in the Dcp2-activating motif on fold-activation of Dcp1–Dcp2 kmax (red), or the KM (blue) determined for each protein complex. Errors are the standard error of individual fits to determine kmax and KM. Primary kinetics data used to construct the plots in (b) and bar graphs in (c) can be found in Supplementary Dataset 1.
