Figure 5.
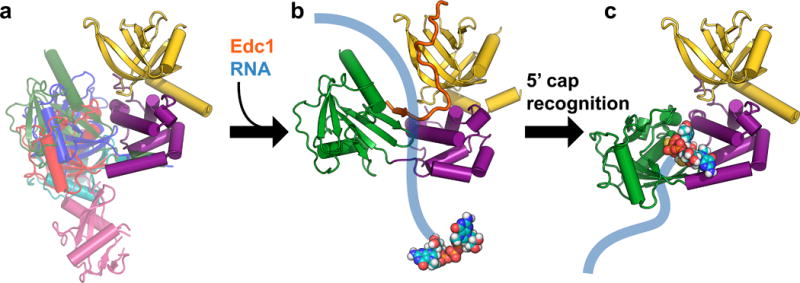
Proposed model for initial steps in the Dcp2 catalytic cycle. (a) The Dcp1–Dcp2 complex exists in an ensemble of conformations in the absence of substrate or coactivators. Dcp1 is colored yellow, the regulatory domain of Dcp2 is colored purple, and different known conformations of the Dcp2 Nudix domain are overlaid in different colors: magenta is the “open” conformation of PDB 2QKM,20 cyan is PDB 2A6T,32 red is PDB 5KQ1 (apo structure from this study; similar to “closed” conformation of PDB 2QKM20), blue is the cap analog-bound conformation from PDB 5KQ4 (this study), and green is the Edc1-bound conformation of PDB 5J3T.39 (b) Coactivators such as Edc1 can bind to the Dcp1–Dcp2 complex and may initially promote a conformation of the apo enzyme that is primed for binding, and possibly subsequent scanning along, RNA. The structure shown in this step is PDB 5J3T,39 with Sp Edc1 coactivator colored orange, and schematic RNA in blue with the 5′ cap shown as spheres. (c) Recognition of the 5′ cap structure on RNA is accompanied by a large conformational change in which the Nudix domain rotates 90° from the Edc1-bound structure shown in (b) to expose previously occluded, conserved cap binding residues to form a composite binding site for cap. The structure shown in this step is PDB 5KQ4, the cap analog-bound conformation from this study. A further conformational change will be required to access the transition state for decapping.
