Figure 2.
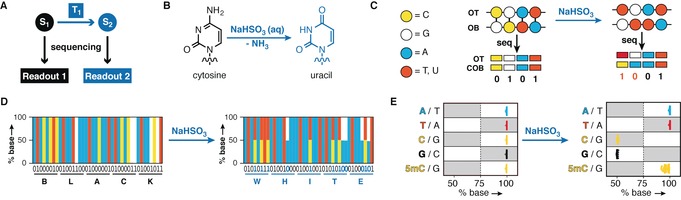
Bisulfite sequencing enables two‐layer encoding in a single nucleic acid template. A) A chemical transformation, T1, can transform an initial state, S1, into a distinct information state, S2. Upon sequencing of both information states, different readouts are obtained. B) Bisulfite mediated cytosine to uracil conversion. C) Deamination of C′s in both the original top (OT) and original bottom (OB) strand with retention of the respective G gives rise to 1:1 mixtures of C:T or G:A reads at these positions after PCR amplification and sequencing (COB=complementary original bottom). Reassigning the binary digits at these positions unravels a second layer of information from a single molecule of DNA. D) Proof‐of‐concept two‐layer encoding by bisulfite‐induced bit conversion. Stacked bar charts display the measured percentage base at a given position across all sequencing reads (same color code applies as for panel C). Shown is a 40 base pair region of an oligonucleotide, which encodes for binary representations of the ASCII text “BLACK” before, and “WHITE” after bisulfite treatment. E) upon bisulfite conversion, a library of oligonucleotides, originally encoding for the first stanza of Edgar A. Poe's The Raven, is read as the second stanza. Sequencing results are presented as percent base calls of 2668 positions. Bisulfite treatment affects 440 C and 420 G positions while 971 A, 789 T, and 68 5mC positions remain unchanged. Gray and white areas denote ranges in which bases are assigned as either 1 or 0 in the reading process. The threshold for calling a bit switch was set at 75 %.
