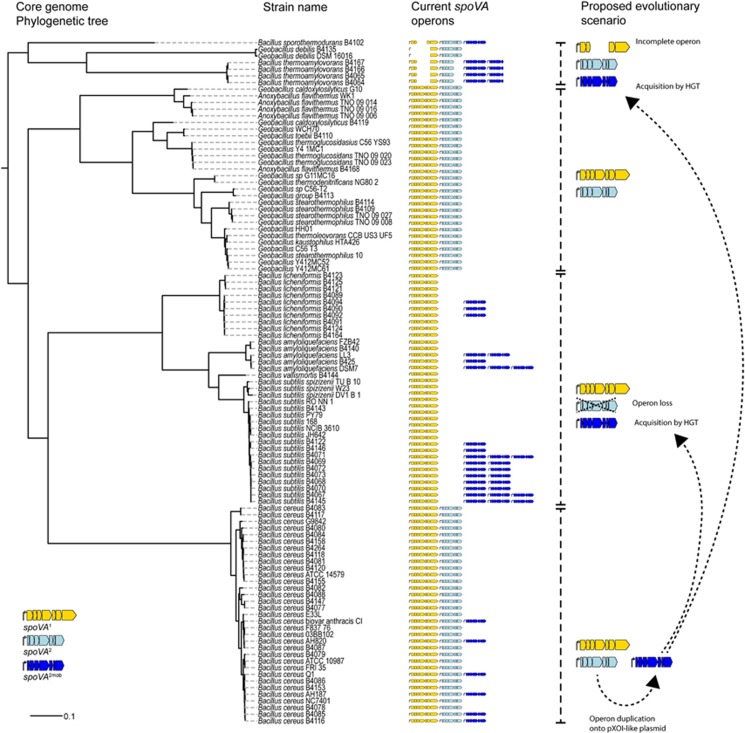
Figure 4.
Maximum likelihood core genome phylogenetic tree of 103 spore-forming Bacillaceae, with indication of the number and type of spoVA operons present in the genomes, and proposed evolutionary scenarios. Three types of spoVA operons were identified in this analysis and are indicated in the tree. First, a spoVA1 operon, encompassing spoVAA, spoVAB, spoVAC1, spoVAD1, spoVAEb1, spoVAEa and spoVAF. Second, a spoVA2 operon, encompassing a gene with a predicted DUF1657 domain, a gene with a YhcN/YljA domain, spoVAC2, spoVAD2, spoVAEb2, a gene with a predicted DUF1657 domain and a gene with a predicted DUF 421 domain and DUF1657 domain. Third, a spoVA2mob operon, which is a duplication of the spoVA2 operon, but present on a mobile genetic element, e.g., Tn1546 in B. subtilis strains. The proposed evolutionary scenarios were based on protein trees of SpoVAC and SpoVAD and the genomic context of the spoVA operons. Strains of B. cereus, Geobacillus spp. and A. flavithermus all carry spoVA1 and spoVA2 operons. Six strains of B. cereus carry spoVA2mob on a pXO1-like plasmid, as part of a Tn1546 transposon. Members of the B. subtilis group (B. subtilis, B. vallismortis, B. amyloliquefaciens, B. licheniformis) lost the spoVA2 operon during evolution, but the spoVA2mob operon re-entered in some strains as part of a Tn1546 transposon. Similarly, spoVA2mob entered strains of B. thermoamylovorans and B. sporothermodurans. Incomplete spoVA1 and spoVA2 operons were observed in strains of B. thermoamylovorans, B. sporothermodurans and C. debilis.