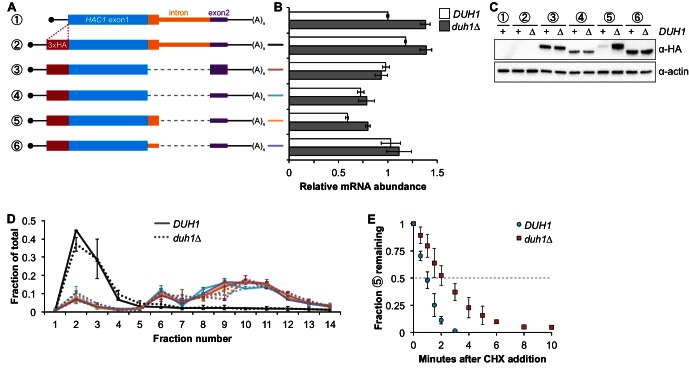
Figure 5. Effects of DUH1 on expression and stability of Hac1p.
(A) Design of 3xHA-tagged HAC1 mRNA variants. Constructs are depicted as in Figure 2B, with the location of the N-terminal 3xHA tag indicated. (B) RNA abundance measurements for HAC1 mRNA variants. Total RNA was extracted from strains expressing the indicated mRNAs in either a DUH1 or duh1∆ background. qRT-PCR was used to measure the abundances of HAC1 variants relative to ACT1 mRNA, with all data normalized to the abundance of construct 1 in strain BY4741. Shown are the mean ± SD (n = 2). (C) Effect of DUH1 disruption on protein abundances. Strains expressing the indicated mRNAs depicted in (A) in either a DUH1 or duh1∆ background were grown to mid-log phase. Extracts were prepared and immunoblotted for 3xHA-Hac1p and actin loading control. (D) Polysome analysis of 3xHA-tagged HAC1 mRNA variants. Extracts were prepared in heparin-containing lysis buffer from strains expressing the mRNAs indicated in (A) in either a DUH1 or duh1∆ background. Polysome analysis was performed as in Figure 1A. (E) Analysis of protein degradation kinetics. Strains expressing construct 5 (depicted in A) in either a DUH1 or duh1∆ background were grown to mid-log phase before being treated with cycloheximide (CHX) to halt translation. At the indicated time points, aliquots of cells were quenched in dry-ice-cold methanol and harvested by centrifugation. Protein extraction and immunoblotting were performed as in (C), except that a high-sensitivity antibody was used to detect 3xHA-Hac1up. Shown are the mean ± SD (n = 3), expressed as a fraction of protein detected at t = 0.