FIGURE 7.
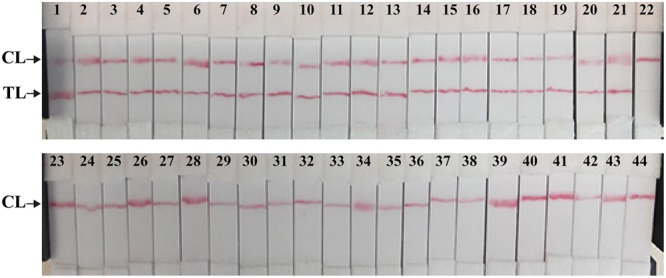
The specificity of MCDA-LFB assay for different strains. The MCDA reactions were conducted using different genomic DNA templates and were monitored by means of visual format. Biosensor 1–18, Shigella flexneri strains of serovar 1d (ICDC-NPS001), 4a (ICDC-NPS002), 5a (ICDC-NPS003), 2b (ICDC-NPS004), 1b (ICDC-NPS005), 3a (ICDC-NPS006), 4av (ICDC-NPS007), 3b (ICDC-NPS008), 5b (ICDC-NPS007), Y (ICDC-NPS010), Yv (ICDC-NPS011), 1a (ICDC-NPS012), X (ICDC-NPS013), Xv (ICDC-NPS014), F6 (ICDC-NPS015), 7b (ICDC-NPS016), 2a1 (ICDC-NPS017), 4b (ICDC-NPS018); biosensor 19–21, Shigella boydii, Shigella sonneri and Shigella dysenteriae; biosensor 22–43, Enteropathogenic E. coli, Enterotoxigenic E. coli, Enteroaggregative E. coli, Enteroinvasive E. coli, Enterohemorrhagic E. coli, Plesiomonas shigelloides, Campylobacter jejuni, Enterobacter cloacae, Enterococcus faecalis, Enterococcus faecium, Yersinia enterocolitica, Streptococcus pneumonia, Aeromonas hydrophil, Vibrio vulnificus, Vibrio fluvialis, Vibrio parahaemolyticus, Klebsiella pneumonia, Bntorobater sakazakii, Bacillus cereus, Listeria grayii, Listeria welshimeri, and Listeria ivanovii; biosensor 44, blank control (DW).
