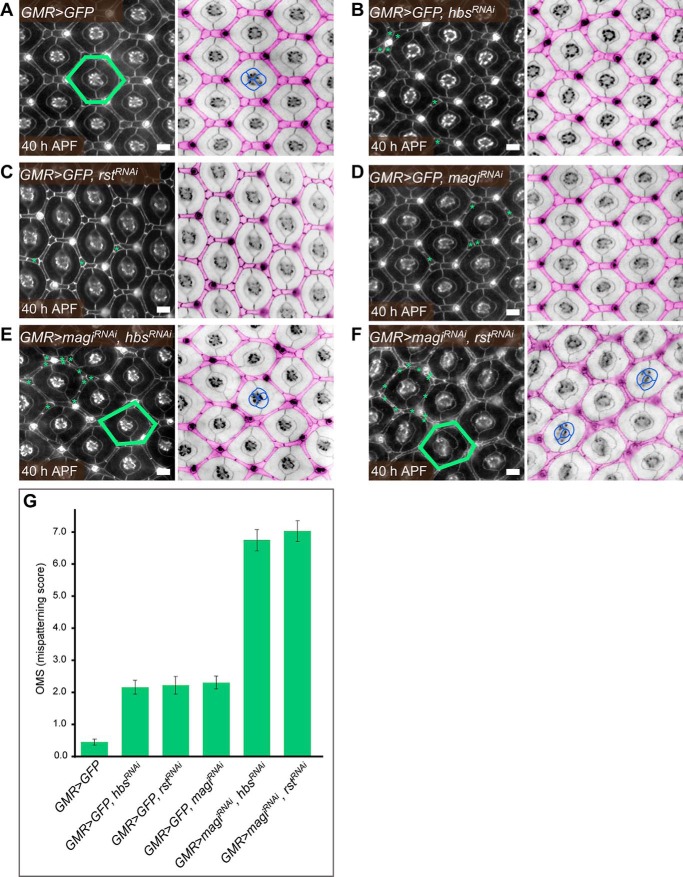
FIGURE 3.
magi, hbs, and rst interact to correctly pattern the Drosophila pupal eye. A, cells were arranged in a wild type patterning when GFP was expressed in the Drosophila pupal eye. In the right-hand panel, the interommatidial cells are pseudocolored in pink. These arranged into stereotypical shapes and positions to generate a hexagonal lattice. One hexagon is indicated in green (left). In the center, four cone cells arranged into a stereotypical pattern (example outlined in blue). The hexagonal pattern of the eye was mildly disrupted when hbs (B), rst (C), or magi (D) was modestly reduced by expression of RNAi transgenes under the control of GMR-Gal4. hbs and rst are fly homologs of mammalian nephrin and neph1, respectively, whereas magi is equivalent to mammalian MAGI-1 or MAGI-2. In each of these genotypes, GFP was co-expressed with the RNAi transgenes. Examples of incorrectly positioned or shaped cells are indicated with green asterisks. Co-expression of magiRNAi and either hbsRNAi (E) or rstRNAi (F) greatly disrupted patterning of the eye. Examples of incorrectly shaped or positioned cells about one ommatidium (a unit eye) are indicated in each left-hand panel. Numerous excess cells were observed. These errors frequently distorted the hexagonal lattice (examples indicated in green). Misarrangements of the four central cone cells were frequent (examples indicated in blue). Scale bars, 10 μm. All eyes were dissected at 40 h after puparium formation (APF). Apical adherens junctions were visualized with an antibody to E-cadherin. G, quantification of the mean number of patterning errors per ommatidium observed in each genotype. Error bars, S.E. See supplemental Table 1 for full statistical analysis.