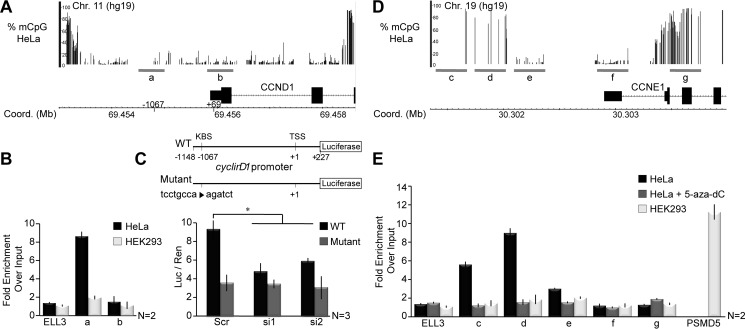
FIGURE 5.
ZBTB33 directly occupies and regulates cyclin D1 and cyclin E1 gene promoters. A and D, WGSBS read coverage tracks depicting percent DNA methylation (mCpG) levels at cyclin D1 and cyclin E1 promoters. Regions within each promoter amplified by ChIP-qPCR are indicated (gray bars). B, ChIP-qPCR analysis at the cyclin D1 promoter in HeLa and HEK293 cells at known ZBTB33 occupation sites, including a KBS-containing location at −1067 and a CpG-containing site at the +69 position. C, scheme depicting design of the wild type (WT) and KBS-mutated minimal cyclin D1 promoter-luciferase reporter plasmids (top). The minimal WT or KBS-mutated cyclin D1 promoter-luciferase reporter plasmids were co-transfected with ZBTBT33 siRNAs, and luciferase activity was quantitated (bottom). Luciferase reads were normalized to Renilla activity. E, ChIP-qPCR analysis for ZBTB33 occupation of the cyclin E1 promoter in HeLa and HEK293 cells at various CpG-containing regions. The PSMD5 gene promoter was selected as a positive control in HEK293 cells based on publicly available ZBTB33 ChIP-seq data (GSM1334009 and GSM803504). Error bars reflect mean ± S.D.; *, p < 0.02 by Student's t test.