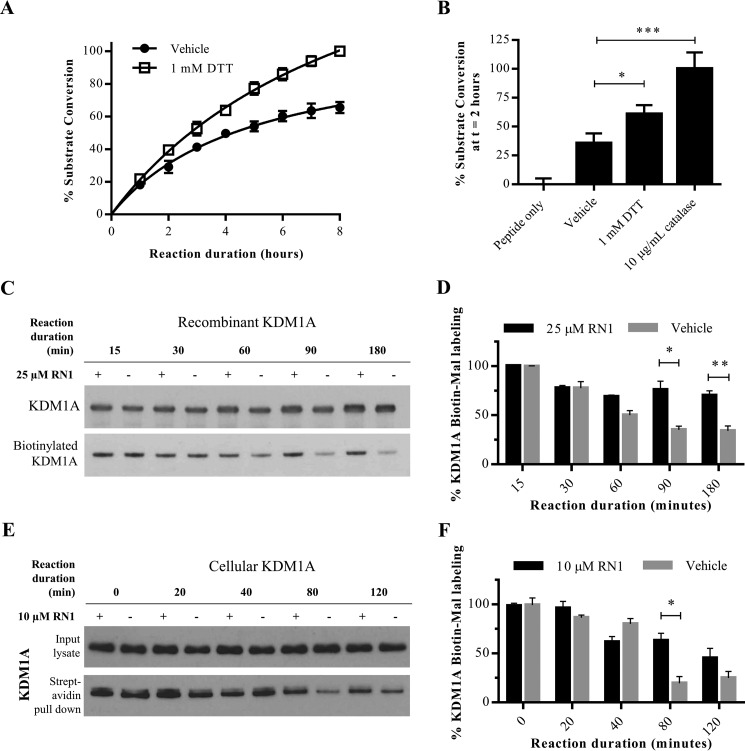
FIGURE 7.
Activity-dependent regulation of KDM1A. A, demethylation of H3K4me2 peptide substrate by recombinant KDM1A is enhanced in the presence of DTT (1 mm), or B, the enzyme catalase (10 μg/ml). For both A and B, demethylation of H3K4me2 starting material was detected by LC-MS. Error bars indicate S.D.; *, p < 0.05; ***, p < 0.001 by one-way ANOVA with correction for multiple comparisons. C, labeling of recombinant KDM1A with Biotin-Mal (25 μm) is reduced with extended demethylation reaction durations. Addition of the FAD-directed inhibitor RN1 (25 μm) blocks the time-dependent reduction in labeling. D, quantification of Biotin-Mal labeling as measured by blotting with a streptavidin-HRP conjugate and normalized to total recombinant KDM1A over three replicate experiments. Error bars indicate S.D.; *, p < 0.05; **, p < 0.01, by 2-tailed t test with correction for multiple comparisons. E, labeling of KDM1A in SH-SY5Y cells by Biotin-Mal (25 μm) is reduced with extended incubation in PBS. Addition of the FAD-directed inhibitor RN1 (25 μm) blocks the time-dependent reduction in labeling. F, quantification of Biotin-Mal labeling as measured by pull down with streptavidin-agarose beads and Western conjugate and normalized to total recombinant KDM1A over three replicate experiments. Error bars indicate S.D.; *, p < 0.05 by 2-tailed t test with correction for multiple comparisons.