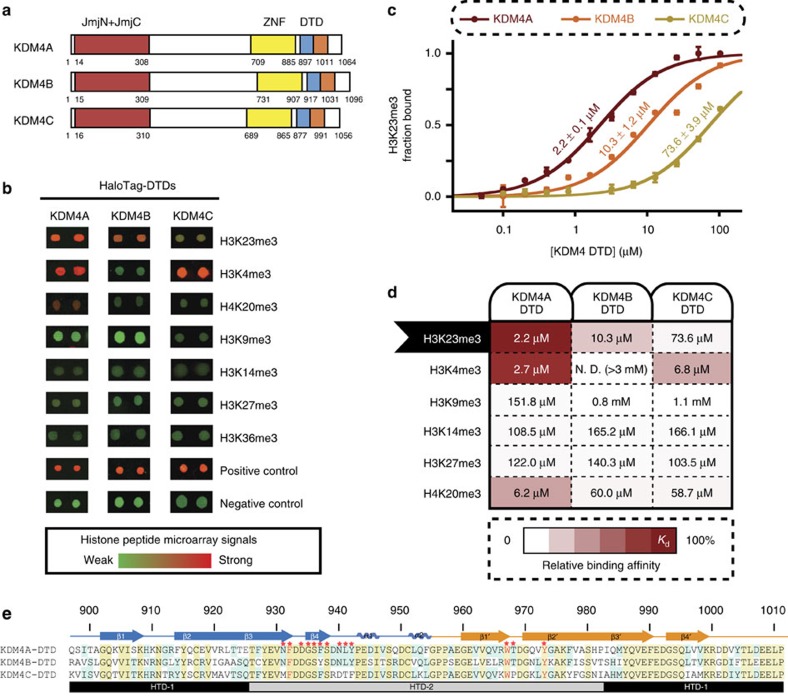
Figure 1. Distinct binding specificities of human KDM4A–C DTDs.
(a) Domain organization of human KDM4 family members (DTD, double tudor domain; JmjN+JmjC, jumonji catalytic demethylase domain; ZNF, zinc finger). DTDs were coloured in two blocks (blue and orange) with each representing a tudor domain sequence. (b) Systematic profiling of histone binding preferences of KDM4A–C DTDs on histone peptide microarray. Recombinant HaloTag-tagged DTDs were expressed and purified form E. coli and incubated with in-house histone peptide microarray. Individual spots were selected from the representative array images (see Supplementary Fig. 1 for the full images). Red signals correlate with binding. (c) Quantitative measurement of H3 (17–32) K23me3 binding constant with KDM4A–C DTDs by fluorescence polarization (FP). Errors represent s.d. from three experimental replicates. (d) Differential histone interactome of KDM4A–C DTDs validated by FP assays. Matrix represents pairs of KDM4 DTD proteins and trimethyl-lysine peptides (see Supplementary Table 1 for peptide information) for each FP assay. Relative specificity factors (as shown by the shades of red) were normalized to the tightest binding in the matrix (Kd=2.2 μM). Binding affinity for each protein-peptide combination as determined by FP is labelled. (e) Sequence alignment of KDM4A–C DTDs. Secondary structure is labelled on top of the sequence (blue: Tudor 1, orange: Tudor 2). Shades indicate conservation of particular residues (yellow: conserved in all three proteins, cyan: conserved in two proteins). Red stars highlight the residues that interact with H3K23me3 peptide as determined from Fig. 2. Letters in red represent aromatic residues.