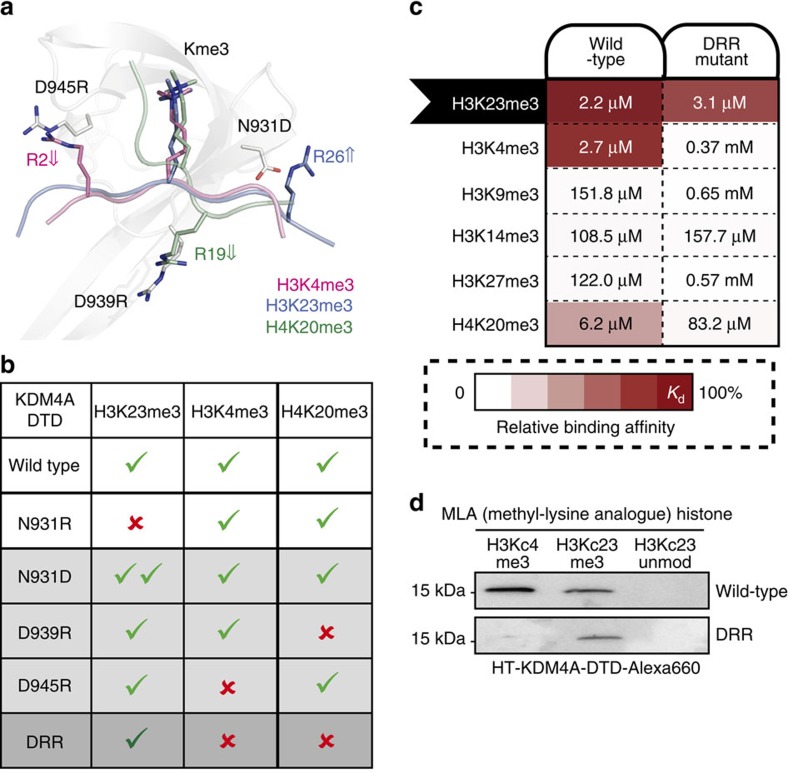
Figure 3. H3K23me3 recognition is modulated by unique side-chain interactions.
(a,b) Structural representation and a table summary about the side-chain interactions for trimethyl-lysine histone recognition by KDM4A-DTD: N931D increases H3R26-mediated H3K23me3 interaction, D939R decreases H4R19-mediated H4K20me3 interaction and D945R decreases H3R2-mediated H3K4me3 interaction. (c) Triple mutant (N931D/D939R/D945R, or DRR) reader is specific for H3K23me3. Matrix represents pairs of KDM4 DTD proteins and trimethyl-lysine peptides (see Supplementary Table 1 for peptide information) for each fluorescence polarization assay. Relative specificity factors (as shown by the shades of red) were normalized to the tightest binding in the matrix (Kd=2.2 μM). Binding affinity for each protein-peptide combination as determined by FP is labelled. (d) DRR mutant fails to recognize H3Kc4me3 MLA but retains binding to H3Kc23me3 MLA histone in a far western blot setting.