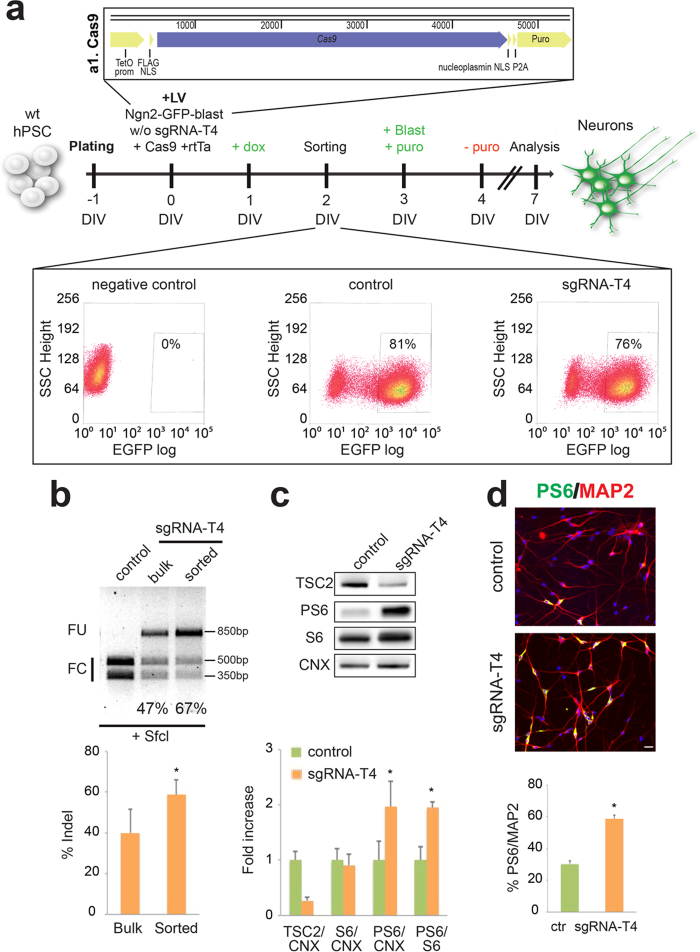
Figure 4. CRISPR/Cas9-mediated TSC2 gene inactivation in wild-type hPSCs.
(a) Top a1, illustration of the Cas9 lentiviral vector expressing Cas9 and puromycin resistance gene (Puro) under the TetO inducible promoter. Middle, schematic representation of the procedure to generate TSC2 mutant neurons starting from wild-type hPSCs. Forced expression of Ngn2 is coupled to TSC2 gene inactivation by co-expressing the sgRNA-T4 guide and Cas9. Doxycycline (added at DIV1) and blasticidin (DIV 3) were maintained for all the experiment. Puromycin was added to select the transduced cells (from DIV3 to DIV4). Bottom, representative FACS plots of EGFP levels in wild-type hPSCs are presented. The lentiviruses used to infect each sample were: Ngn2-blast for the negative control, Ngn2-GFP-blast for the control and Ngn2-GFP-blast-sgRNA-T4 for the sgRNA-T4 sample. The EGFP positive population selected for the study is enclosed in the box area. (b) Representative SfcI RFLP assay performed in control sorted, sgRNA-T4 bulk and sgRNA-T4 sorted populations derived from wild-type hPSCs. Quantification of indel percentage by RFLP in sgRNA-T4 bulk and sgRNA-T4 sorted cells (n = 4, *p < 0.05). (c) Western-blots to detect TSC2, PS6, S6 and Calnexin (CNX) protein levels in control and sgRNA-T4 neurons. Densitometric quantification of TSC2, PS6 and S6 relative to CNX or S6 protein levels in arbitrary units (n = 3, *p < 0.05). (d) Immunostaining of the sorted cells for MAP2 and PS6. Quantification of PS6/MAP2 double positive cells respect to the total MAP2-positive cell population (n = 3, *p < 0.05). Nuclei were stained with Hoechst. Scale bar: 20 μm. Full-length gel and blots are included in Figure S7e and f.