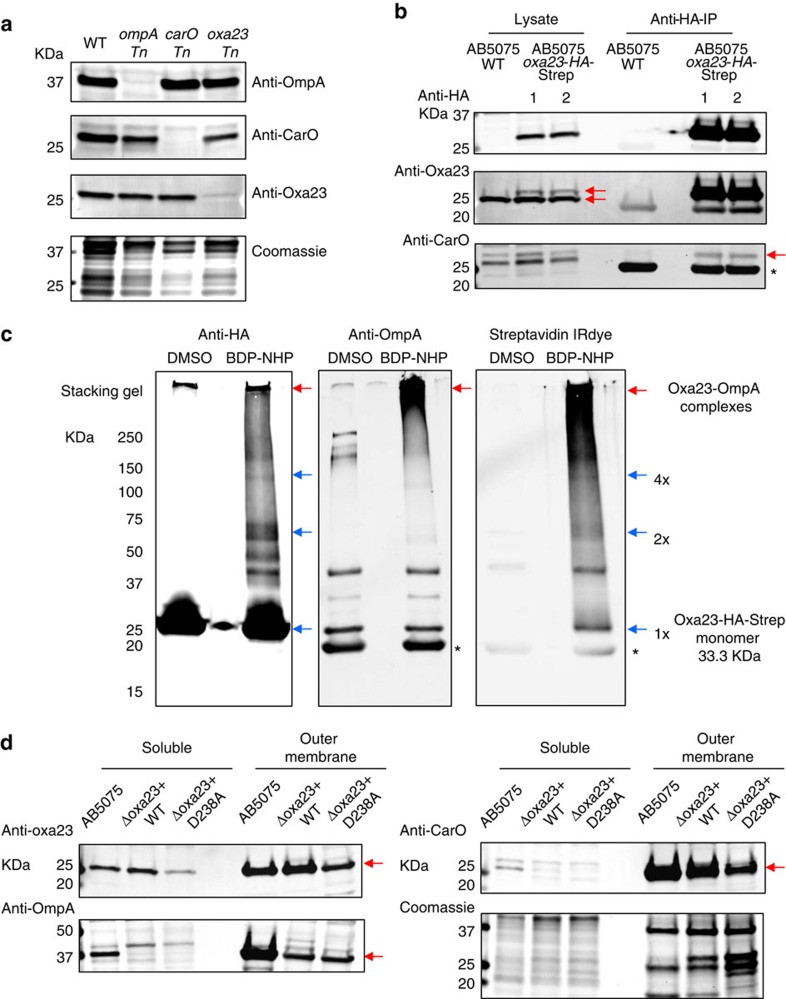
Figure 4. Validation of Oxa-23 interactions with CarO and OmpA.
(a) Antibody specificity was confirmed with detection of protein bands that matched the molecular weight of the target proteins (OmpA, CarO and Oxa-23). Furthermore, the detected protein bands also matched the sizes of protein bands that showed reduction in specific transposon mutants (ompA Tn, carO Tn and oxa-23 Tn). (b) Co-IP analysis of anti-HA antibodies with AB5075-pMMB-oxa23 cell protein lysates. Red arrows in anti-Oxa23 immunoblots indicate the detection of both the endogenous Oxa-23 and the tagged Oxa-23. Red arrow in anti-CarO immunoblots indicates CarO proteins that were co-immunoprecipitated with Oxa23-HA-Strep in AB5075. Asterisk corresponds to the immunoglobulin light chains. (c) Co-IP analysis of anti-HA antibodies with the BDP-NHP cross-linked AB5075-pMMB-oxa23 cells. Blue arrows indicate that protein bands matching the sizes of Oxa23-HA-Strep monomers (1 × ), dimers (2 × ) and tetramers (4 × ) could be immunoprecipitated with anti-HA antibodies after the BDP-NHP cross-linking reactions. Red arrows show that higher molecular weight bands reactive to anti-OmpA antibodies could be pulled down with the anti-HA antibodies, supporting the identification of Oxa23-OmpA protein interactions. Streptavidin IRdye confirmed that the high molecular weight bands contained strong biotin (that is, cross-linking) signals in the cross-linked samples, but not in the dimethyl sulfoxide (DMSO)-treated control samples. Asterisks correspond to the immunoglobulin light chains. (d) OM protein-enriched fractions were prepared from AB5075 WT, or Δoxa23 complementation mutants with pMMB.A1-oxa23 (−WT or −D238A) cells. Two micrograms of protein were loaded for each lane. Strong anti-Oxa23 immunoblot signals were observed at the OM-enriched fractions. Enrichment of OM-protein markers OmpA and CarO are also shown in these samples. Red arrows indicate the detection of specific protein bands, Oxa-23, OmpA and CarO. Coomassie-stained gel shows the loading of equal total proteins. Full blots are shown in Supplementary Fig. 15.