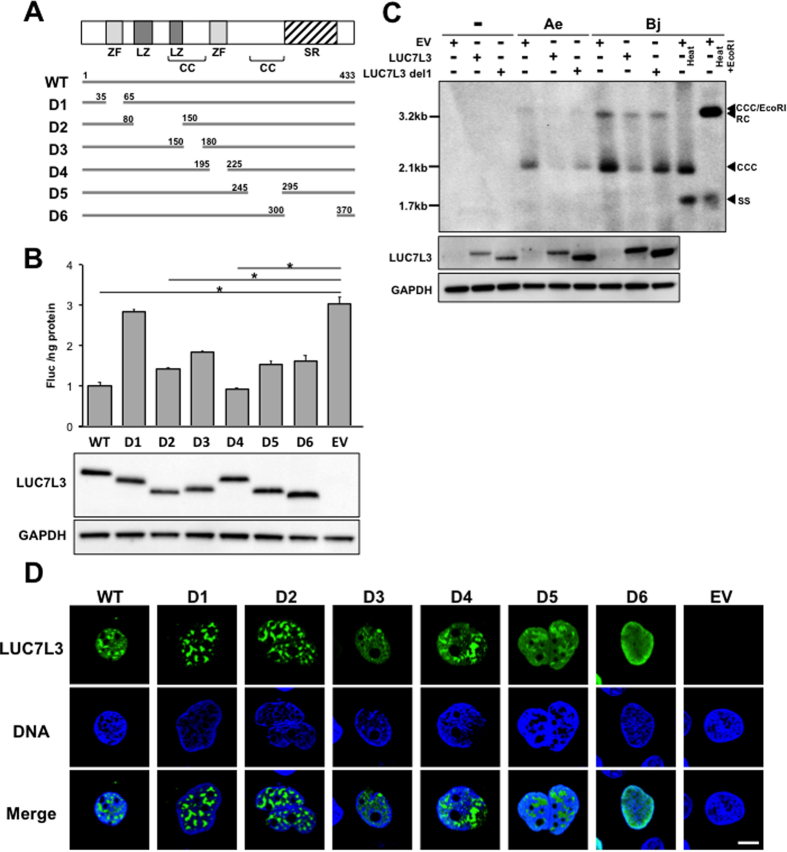
Figure 3. Effects of deletions within LUC7L3 on its subcellular distribution and on expression of the viral pgRNA.
(A) A series of deletion mutants of LUC7L3 used in this study were indicated. A schematic diagram of predicted domain structures of LUC7L3 is indicated at the top. ZF, zinc-finger domain; LZ, leucine zipper domain; SR, SR-rich domain; CC, coiled-coil domain. (B) HuH7 cells were transfected with FLAG-tagged LUC7L3 (LUC7L3) or its partial deletions (D1–D6) or empty vector (EV) together with pGLHBp1627/1817 (1627/1817). Cells were harvested three days later to determine the luciferase activities (upper) and expression of LUC7L3 proteins as well as of GAPDH was assessed by immunoblotting (lower). (C) HuH7 cells were transfected with FLAG-tagged LUC7L3 (LUC7L3) or its partial deletion (LUC7L3 D1) or empty vector (EV) together with pUC-HBAe, -Bj or pUC19 (−). At 5 days post-transfection, Hirt DNA was prepared from the transfected cells and subjected to Southern blot analysis. RC, rcDNA; CCC, cccDNA; SS, single-strand DNA. cccDNA was resistant to denaturation at 85 °C for 5 min (Heat), but sensitive to EcoRI digestion (Heat + EcoRI). (D) Subcellular localization of LUC7L3 and its deletion mutants were determined by confocal microscopy images of cells transfected with expression plasmids for FLAG-tagged LUC7L3 (LUC7L3) or its partial deletions (D1–D6) or empty vector (EV). Cells were immunostained with anti-FLAG antibody as a first antibody (LUC7L3), and counterstained with Hoechst 33342 to label nuclei (DNA). Scale bar, 10 μm. Statistical significances compared with EV were shown. *p < 0.05, Student’s t test.