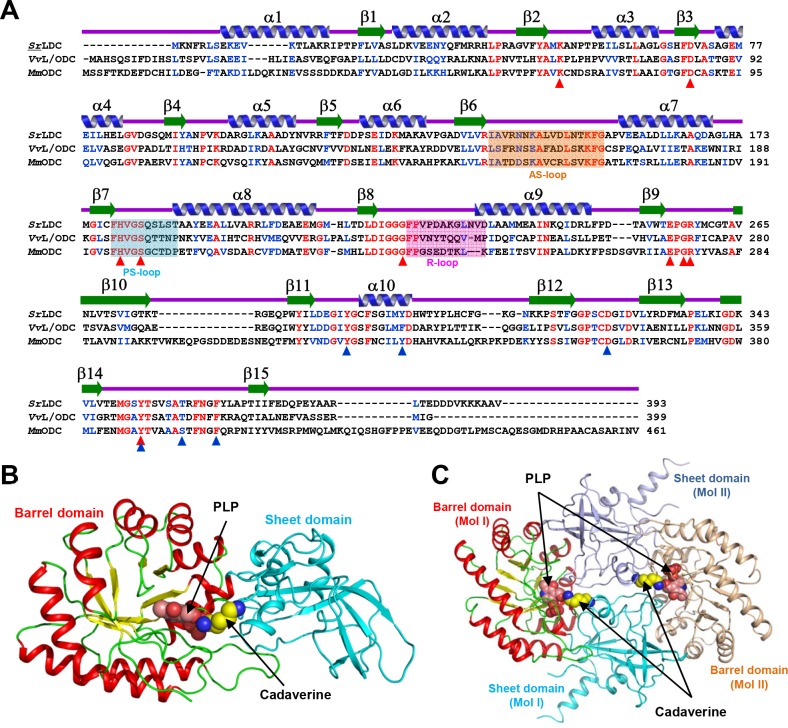
Fig 1. Crystal structure of SrLDC.
(A) Amino acid sequence alignment of L/ODCs. Identical and highly conserved residues are presented red and blue colored characters, respectively. Secondary structure elements are drawn based on the SrLDC structure in an apo-form. The AS-loop, PS-loop and R-loop are shown in boxes with orange, cyan and magenta colors, respectively. Residues involved in binding of PLP and substrate are marked with red- and blue-colored triangles, respectively. SrLDC, VvL/ODC, and MmODC are representatives of lysine decarboxylase from Selenomonas ruminanitum, lysine/ornithine decarboxylase from Vibrio vulnificus, and ornithine decarboxylase from Mus musculus, respectively. (B) Monomeric structure of SrLDC. A monomeric protein is shown as a cartoon diagram. The barrel domain is presented with colors of red and yellow for α-helices and β-strands, and the sheet domain is with a cyan color. The bound PLP and cadaverine are shown as sphere models with salmon and yellow colors, respectively. (C) Dimeric structure of SrLDC. The dimeric structure of SrLDC is shown as a cartoon diagram showing one monomer with a color scheme in (B) and the other monomer in light-orange and light-blue for the barrel domain and the sheet domain, respectively. The bound PLP and cadaverine are shown as sphere models with salmon and yellow colors, respectively.