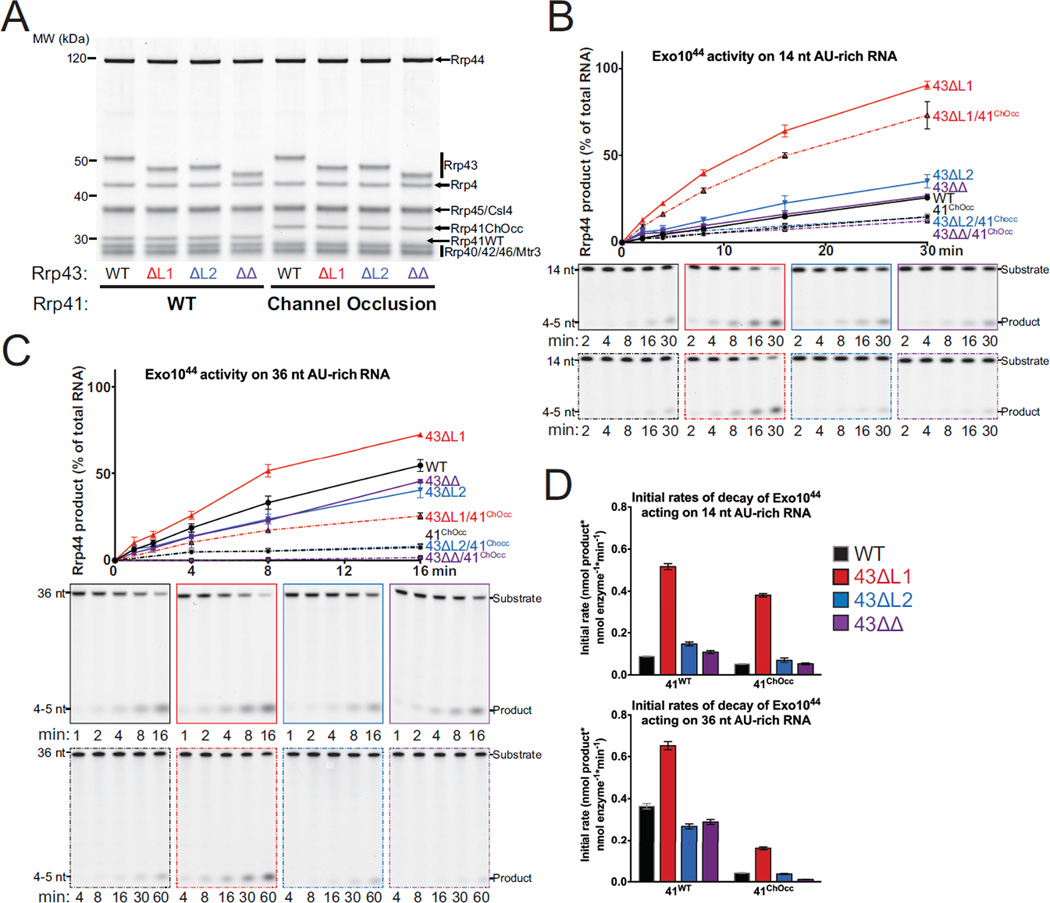
Figure 3. Exo1044 Activity on AU-rich RNAs is Enhanced by Deletion of Rrp43L1.
(A) SDS-PAGE analysis of reconstituted Exo1044 complexes containing Rrp43 mutants. ‘41ChOcc’ refers to a loop insertion mutant in Rrp41 that occludes the central channel.
(B and C) WT and mutant Exo1044 activity on 14 nt (B) and 36 nt (C) 5’ FAM AU-rich RNA substrates. The experiment was performed in triplicate and quantification of mean values is shown with error bars at ±1 standard deviation. Representative polyacrylamide TBE-urea gels are shown below.
(D) Quantification of initial rates of decay of Exo1044 complexes on 14 nt and 36 nt 5’ FAM AU-rich RNA substrates from the experiments in panels (B) and (C). Mean values of the triplicate experiments are plotted with error bars at ±1 standard deviation. See also Figure S4.