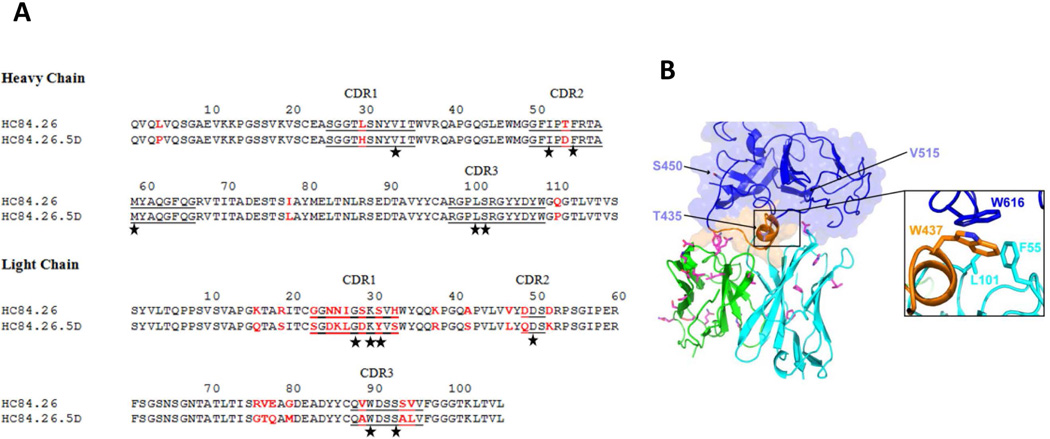
Figure 4. Affinity-matured residues in the HC84.26.5D-E2434–446 interface and model of HC84.26.5D with E2 core protein.
(A) Amino acid sequences of the VH and VL regions of wild-type HC84.26 and affinity-matured HC84.26.5D were aligned. Bars show the position of CDRs. Amino acid differences between the two antibodies are highlighted in red. Contacting residues in the HC84.26.5D–E2434–446 structure are marked with black stars. (B) Model of HC84.26.5D in complex with E2 core glycoprotein. VH and VL domains are cyan and green, respectively. Mutant antibody residues from affinity maturation are drawn in stick format, with carbon atoms in magenta, nitrogen atoms in blue, and oxygen atoms in red. Three sites that were mutated in the HC84.26.5D-treated mouse with breakthrough infection (B.A818), and not inoculum or control mice, are shown as light blue sticks on the E2 core model and labeled. Inset shows putative antibody VH hydrophobic contacts with E2 residue W616 based on the modeled complex (E2 residue W437 also shown for reference).