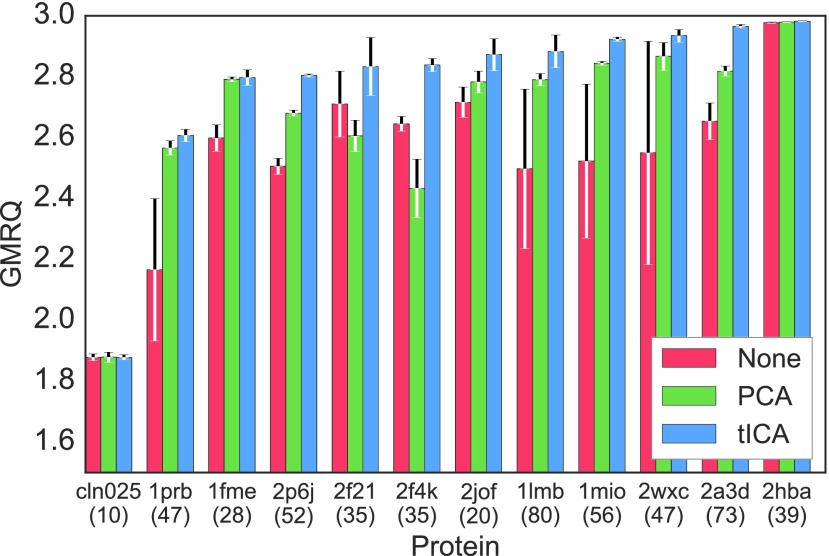
FIG. 3.
GMRQ scores for two-time scale MSMs generated for twelve ultra-long protein folding datasets18 where additional dimensionality reduction has been omitted (red), performed with PCA (green) or performed with tICA (blue) demonstrate that tICA systematically improves models. All MSMs were made using α-angle featurization and mini-batch k-means clustering. In all cases, the best tICA model performs better than or equivalently to both the best PCA model and the best model created directly from α-angle features. The error bars signify the score standard deviation generated from five cross-validation iterations. The number of amino acids of each protein is given below its PDB ID.