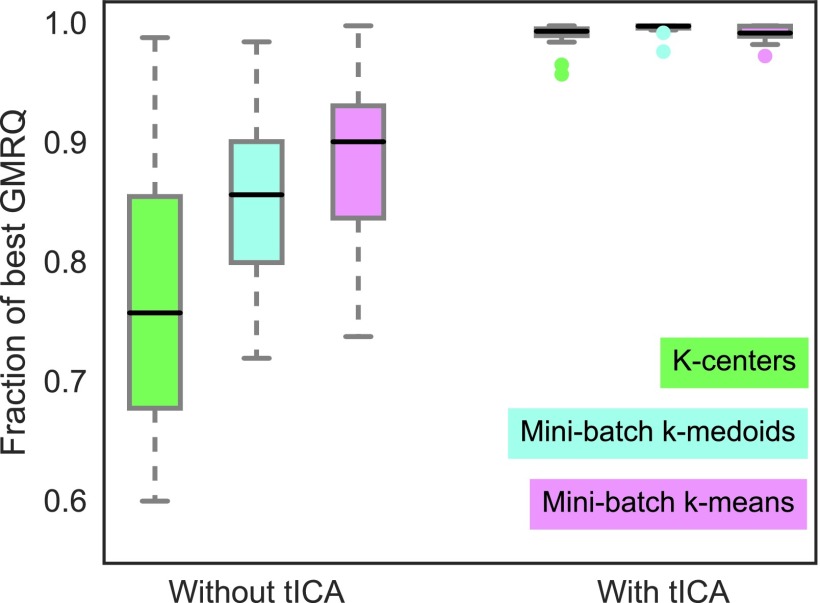
FIG. 4.
Aggregated score ratios for two-time scale MSMs generated for twelve ultra-long protein datasets18 using three different clustering algorithms with or without tICA show that different clustering algorithms produce similarly well-performing models when tICA is used. All models were made using α-carbon contact distances. When clustering is performed directly from features, the dimensionality is reduced by 2-3 orders of magnitude; whereas when clustering is performed from tICs, dimensionality is reduced from or lower to . For large dimensionality reductions, k-means clustering produces the best models. For small dimensionality reductions via tICA, clustering algorithms produce similarly well-performing models that are categorically better than models created without tICA. (The best-performing algorithm at small dimensionality reductions is k-medoids; see the supplementary material, Fig. S9.)