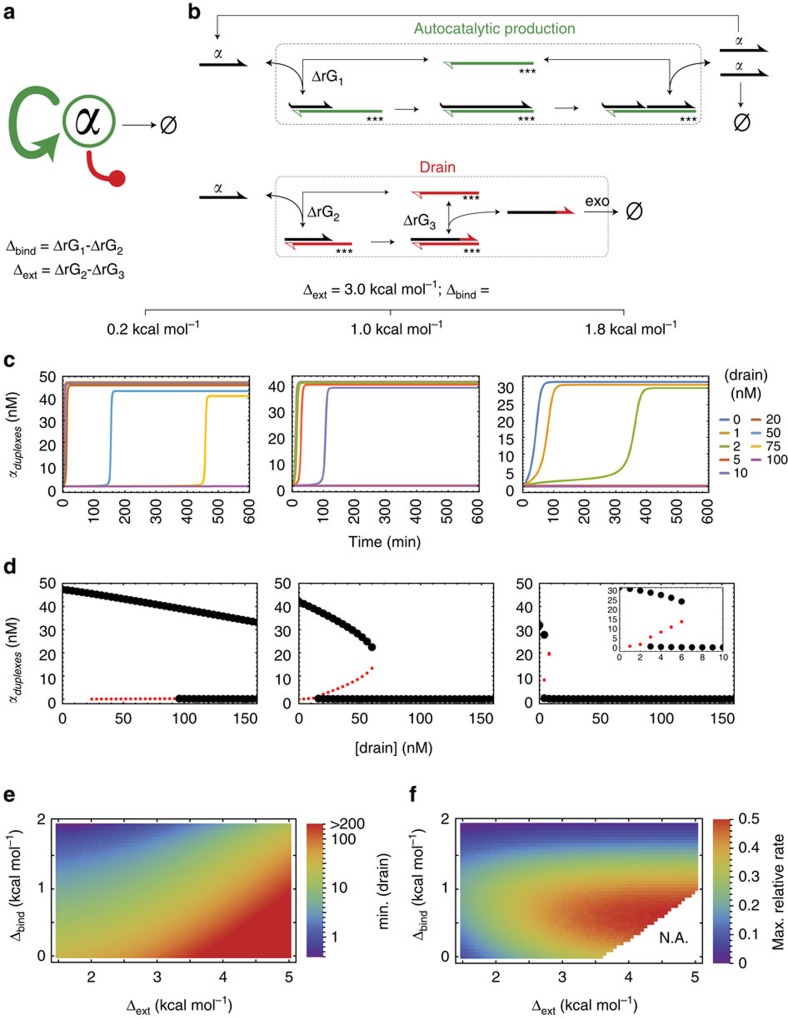
Figure 2. Drain-based bistability design for the PEN DNA toolbox.
(a) Schematic representation of a simple autocatalytic network with a sink and an additional decay pathway (dead-ended red arrow). The concentration of α is the dynamic variable considered here. (b) The species-specific and saturable degradation pathway is constructed from a short drain template that is able to bind the input α, extend it with a few additional bases and slowly release this deactivated species for eventual degradation. The difference between the binding strength on the drain and on the replication template (Δbind) controls the respective reaction fluxes at low α concentration. The maximum throughput of the drain pathway depends on the dehybridization step, hence it is governed by the stabilization of the trigger upon extension on the drain (Δext). Empty arrowheads show non-extensible 3′-ends (3′ phosphate modification), and *** denotes the protection against exonuclease degradation (backbone modification). (c,d) Predicted time traces and fixed points (stable in black, unstable in red), computed at Δext=3.0 kcal mol−1 using a complete kinetic model. The y axis corresponds to the total concentration of complexes involving α. Curves suggest that the bistable behaviour can be observed for a variety of Δbind values, if enough drain is present. Higher Δbind require less drain, but lead to sluggish amplifications and quickly fall back in a monostable trivial behaviour. (e) Computed concentrations of drain template necessary to reach the bistable regime in the Δbind/Δext parameter space (taken to reflect 0–3 deletions on the template and 2–6 bases on the drain tail). (f) Computed maximum amplification rates compatible with bistability (that is, at the minimal drain concentration). Values corresponding to drain concentrations above 200 nM were not computed.