Figure 4.
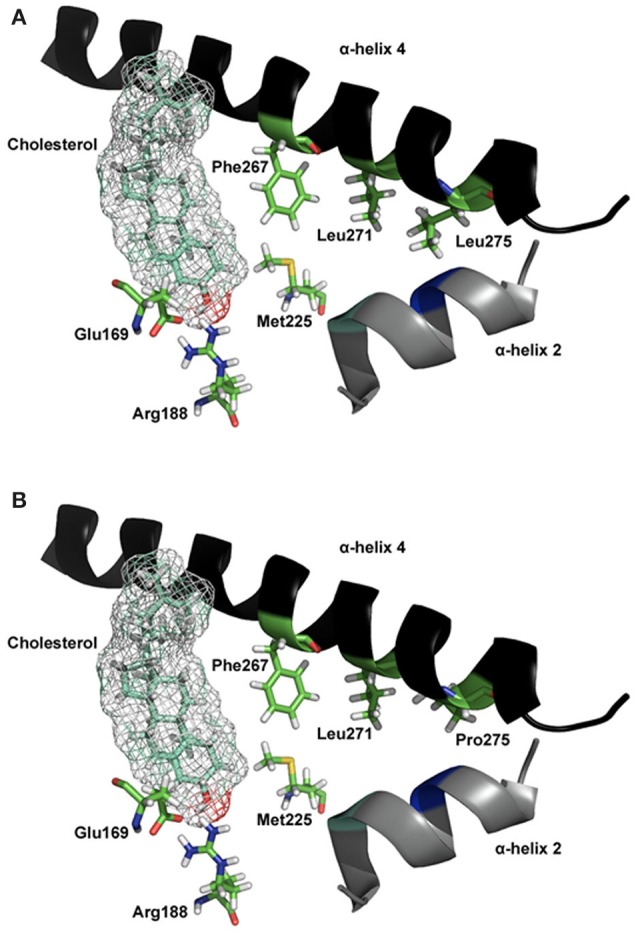
Molecular model of STARD1 L275P mutation. (A) The essential elements of the wild-type STARD1 are shown. The salt bridge formed by Glu169/Arg188 putatively interacts with the β-OH of cholesterol, while the α-helix 4 is in a closed state. The line of hydrophobic residues Phe267/Leu271/Leu275 interacting with α-helix 2, Met225 and other hydrophobic residues (not shown for image simplicity) help stabilize the STARD1 /cholesterol complex. (B) The long hydrophobic side chain for Leu275 is absent in the clinical mutation L275P, thereby creates a void in the hydrophobic environment necessary for stabilizing holo- STARD1, leading to a reduction in cholesterol binding and steroidogenic activity (Roostaee et al., 2008). Initial coordinates for the molecular modeling of STARD1 were retrieved from the Protein Databank (code 1IMG). Site-directed mutagenesis and energy minimization were done in silico using the molecular modeling software SYBYL 8.0 (Tripos Inc, St. Louis, MO). All the rendering was done using PYMOL (DeLano, W. L. The PyMOL Molecular Graphics System, 2002, DeLano Scientific, Palo Alto, CA).
