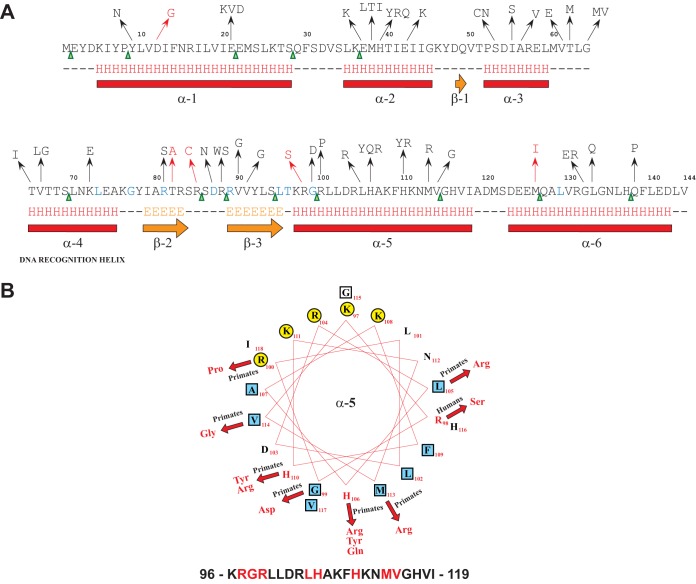
FIG 1.
Location of nonsynonymous substitutions within FabT. (A) Secondary-structure prediction for FabT derived from Jpred. Nonsynonymous substitutions are represented in black (mutations identified in isolates cultured from nonhuman primates) and red (mutations identified in isolates cultured from humans). Stop codons are represented by green arrowheads (mutations identified in isolates cultured from nonhuman primates). The residues in blue are conserved among members of the MarR superfamily of proteins. (B) Helical-wheel representation of the FabT amphipathic alpha helix 5 (α5). The amino acid sequence, with the mutated residues in red, is shown at the bottom; polar and hydrophobic residues are represented by yellow circles and blue squares, respectively. The resulting nonsynonymous substitutions are shown in red, and the arrows denote the corresponding changes. Primates, nonhuman primates; Humans, host from which the respective mutated GAS strain was isolated.