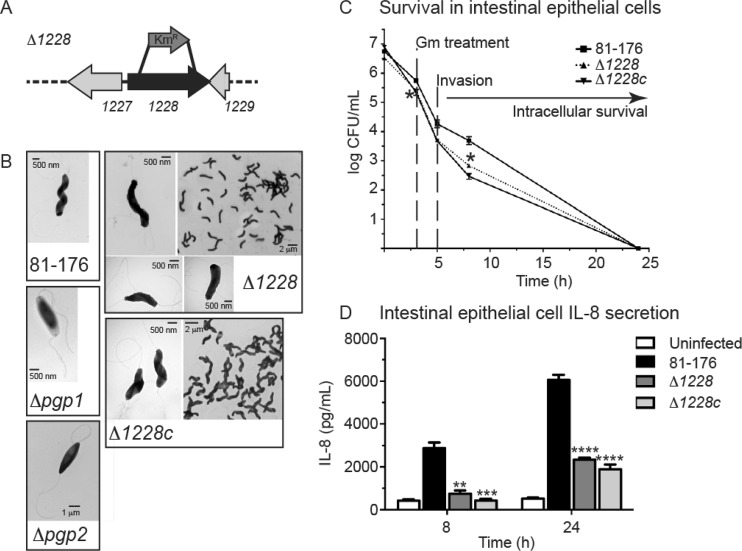
FIG 1.
C. jejuni cjj81176_1228 (1228) gene locus and morphology, invasion and intracellular survival, and IL-8 secretion levels of the Δ1228 strain. (A) Genomic organization of cjj81176_1228 (denoted as 1228) gene locus. The Δ1228 mutant strain was constructed by deleting 1,014 bp of the 1,161-bp gene and replacing it with the nonpolar aphA3 Kmr cassette. (B) Negatively stained transmission electron microscopy images of the helical C. jejuni 81-176 strain, the rod-shaped Δpgp1 (14) and Δpgp2 (15) mutant strains, C- and S-shaped Δ1228 mutant strain, and Δ1228 complemented strain (Δ1228c) with restored helical morphology. All strains show intact flagella. (C) Invasion by and intracellular survival of the C. jejuni wild type 81-176 Δ1228 mutant and Δ1228 complemented strain (Δ1228c) in the INT407 epithelial cell line were assessed by the gentamicin (Gm) protection assay. The Δ1228 mutant strain showed a slight defect in invasion and intracellular survival. Gm was added 3 h postinfection. After 2 h, the Gm was washed off and the cells were incubated with fresh MEM containing 3% fetal bovine serum and a low dose of Gm. At each time point, CFU were determined for each well by lysing the cells with water and plating the dilutions onto MH-TV plates. Data represent the mean ± standard error of the mean from three replicates and are representative of three independent experiments. (D) ELISA was used to quantify IL-8 levels secreted by uninfected INT407 epithelial cell lines and cells infected for 8 and 24 h with C. jejuni wild-type 81-176, Δ1228, and Δ1228c strains. The Δ1228 mutant resulted in decreased IL-8 secretion in comparison to wild type. Data represent the mean ± standard error of the mean from three replicates and are representative of three independent experiments. Asterisks indicate a statistically significant difference using the unpaired Student t test, with *, **, ***, and **** indicating P < 0.05, P < 0.01, P < 0.001, and P < 0.0001, respectively.