Figure 3.
Rbins Inhibit Mdn1’s ATPase Activity In Vitro
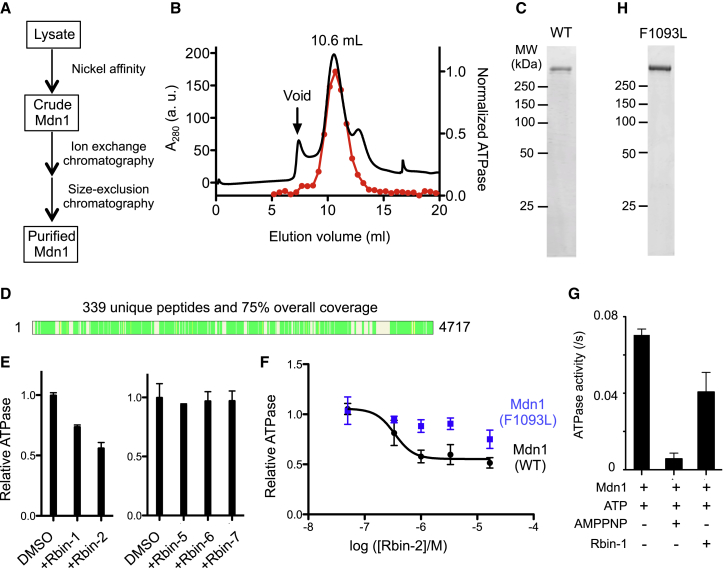
(A) Schematic for strategy used to purify recombinant full-length Mdn1.
(B) The size-exclusion chromatography profile for Mdn1 (black trace). The activity of different fractions from this chromatography step was analyzed using a radioactive ATPase assay ([MgATP] = 0.1 mM). Activity was normalized to the most active fraction (red dots, each fraction; trace, interpolation).
(C) SDS-PAGE analysis of purified full-length Mdn1 (WT) (Coomassie stain).
(D) Peptides identified by mass spectrometry of the purified protein are indicated (green bars, schematic generated using Proteome Discoverer 1.4, Thermo Scientific).
(E) Mdn1’s activity in the presence of Rbin analogs (1 μM) was tested using an NADH-coupled ATPase assay ([MgATP] = 1 mM). The relative activity is indicated (mean ± range, n = 2 independent experiments).
(F) Dose-dependent inhibition of the steady-state ATPase activity of wild-type Mdn1 and Mdn1(F1093L) by Rbin-2 (mean ± SD, n = 4 independent experiments). An apparent EC50 was estimated for the inhibition of wild-type Mdn1 using a sigmoidal dose-response curve. Using similar equations, we were unable to properly fit the small decrease in activity of Mdn1(F1093L) across the concentration range tested. Unpaired t test (without Welch’s correction) of the measured activity for WT and Mdn1(F1093), at the highest three inhibitor concentrations tested, indicate that the difference in values are statistically significant (p = 0.0020 (at 16.7 μM), p = 0.0021 (at 3.33 μM), and p = 0.0019 (at 1.0 μM)).
(G) The ATPase activity of Mdn1 in the presence Rbin-1 (1 μM) or AMP-PNP (2 mM) using the radioactive ATPase assay (MgATP = 0.1 mM). Error bars show SD (n = 6 independent experiments).
(H) SDS-PAGE analysis of purified full-length Mdn1(F1093L) (Coomassie stain).
See also Figure S4.