Abstract
The Gram-negative bacterial pathogen Pseudomonas aeruginosa uses three interconnected intercellular signaling systems regulated by the transcription factors LasR, RhlR, and MvfR (PqsR), which mediate bacterial cell–cell communication via small-molecule natural products and control the production of a variety of virulence factors. The MvfR system is activated by and controls the biosynthesis of the quinolone quorum sensing factors HHQ and PQS. A key step in the biosynthesis of these quinolones is catalyzed by the anthranilyl-CoA synthetase PqsA. To develop inhibitors of PqsA as novel potential antivirulence antibiotics, we report herein the design and synthesis of sulfonyladeonsine-based mimics of the anthranilyl-AMP reaction intermediate that is bound tightly by PqsA. Biochemical, microbiological, and pharmacological studies identified two potent PqsA inhibitors, anthranilyl-AMS (1) and anthranilyl-AMSN (2), that decreased HHQ and PQS production in P. aeruginosa strain PA14. However, these compounds did not inhibit production of the virulence factor pyocyanin. Moreover, they exhibited limited bacterial penetration in compound accumulation studies. This work provides the most potent PqsA inhibitors reported to date and sets the stage for future efforts to develop analogues with improved cellular activity to investigate further the complex relationships between quinolone biosynthesis and virulence factor production in P. aeruginosa and the therapeutic potential of targeting PqsA.
Pseudomonas aeruginosa is an opportunistic, Gram-negative bacterial pathogen that poses a serious threat in nosocomial infections, particularly in immunocompromised patients, such as burn victims, those undergoing cancer therapy, and HIV-infected individuals.1 It is also a leading cause of death in cystic fibrosis patients.2P. aeruginosa is highly prone to antibiotic resistance through both intrinsic mechanisms such as restricted cell permeability, antibiotic efflux, and biofilm formation as well as acquired mechanisms such as target mutation and enzymatic drug inactivation via horizontal gene transfer.3,4 A promising strategy to overcome this growing and challenging resistance problem in P. aeruginosa is to target nonvital functions that are associated with pathogenicity, such as the production of virulence factors.5,6 Because virulence factors are not directly associated with bacterial viability, antivirulence antibiotics may be less susceptible to the development of drug resistance.7,8
The synthesis and secretion of numerous virulence factors in P. aeruginosa are controlled by three quorum sensing systems that mediate bacterial cell–cell communication using small-molecule natural products.9,10 The LasR- and RhlR-regulated systems use acyl homoserine lactones (AHL) as signaling molecules, while the MvfR (PqsR)-regulated system uses two quinolones, PQS (3,4-dihydroxy-2-heptylquinoline, Pseudomonas Quinolone Signal)11 and its biosynthetic precursor HHQ (2-heptyl-4-hydroxyquinoline).12 By binding the transcriptional activator MvfR (PqsR), PQS and HHQ induce the expression of a variety of virulence factor genes including their own biosynthetic genes, promote biofilm formation, and interact with the distinct AHL-based quorum sensing systems.12−18 Although both HHQ and PQS bind to and activate MvfR, PQS is 100-fold more potent than HHQ.12 Inhibition of HHQ and PQS biosynthesis should therefore prevent MvfR activation, and consequently MvfR-dependent gene regulation. Toward this end, we report herein the design, synthesis, and biochemical, cellular, and pharmacological evaluation of small-molecule inhibitors of PqsA, an acyl-CoA synthetase used in quinolone biosynthesis. Using rational design based on enzyme mechanism, we have developed potent inhibitors of PqsA that block quinolone biosynthesis in P. aeruginosa.
Results and Discussion
Design and Synthesis of PqsA Inhibitors
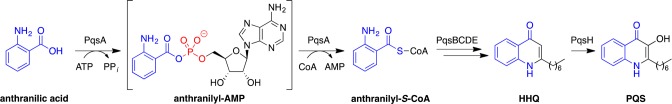
Biosynthesis of the quinolone HHQ is thought to involve PqsA–E, with final conversion to PQS involving oxidation by PqsH (Figure 1).19,20 Inhibitors of the cyclase PqsD22−29 and the thioesterase activity of PqsE,30 direct antagonists of the MvfR receptor,31−37 a dual inhibitor of PqsD and MvfR,38 as well as an inhibitor of the upstream enzyme KynU, which is one source of the anthranilic acid precursor,39 have been reported. Our efforts to target this pathway have focused on PqsA, an acyl-CoA synthetase (ligase) that catalyzes a key step in quinolone biosynthesis.21 PqsA converts anthranilic acid to anthranilyl-CoA via a two step process involving initial ATP-dependent adenylation of anthranilic acid to form a tightly bound anthranilyl-AMP intermediate, followed by thioesterification with CoA to form anthranilyl-CoA. PqsA has been validated previously as a promising therapeutic target using both genetic and pharmacological approaches. pqsA– mutants do not produce HHQ or PQS15,40 and are poor biofilm producers in vitro.41 They also exhibit attenuated virulence in a mouse burn infection model,16,42 reduced dissemination in a mouse lung infection model,40 and less biofilm formation and increased susceptibility to clearance by ciprofloxacin in a mouse tumor infection model.43 There is no known human orthologue of PqsA, although related aliphatic acyl-CoA synthetases are used in metabolism. Thus, selective PqsA inhibitors would not be expected to impact host cell viability. Moreover, haloanthranilate substrate analogues that act as either PqsA substrates or inhibitors have been shown to inhibit PQS production in cell culture21 and to decrease both quinolone production and mortality in the mouse burn model.42
Figure 1.
Biosynthesis of the P. aeruginosa quinolone quorum sensing factors HHQ and PQS is initiated by PqsA, an anthranilyl-CoA synthetase that first activates anthranilic acid to form a tightly bound anthranilyl-AMP reaction intermediate and then catalyzes thioesterification with CoA to form anthranilyl-CoA.
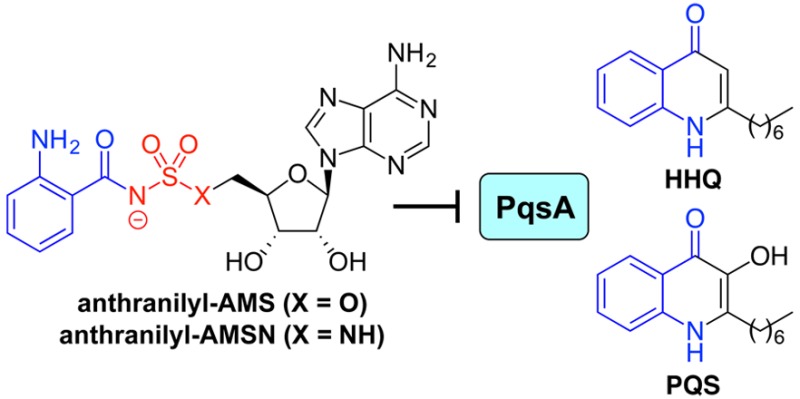
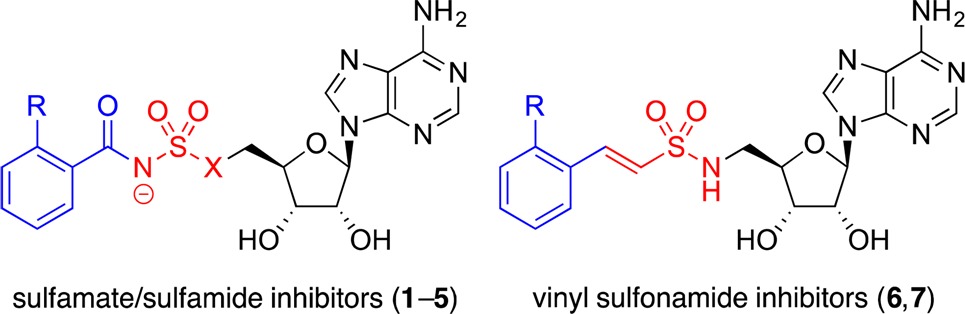
Acyl-CoA synthetases belong to a superfamily of structurally and mechanistically related adenylate-forming enzymes that also includes nonribosomal peptide synthetase (NRPS) adenylation domains and firefly luciferase.44 We45−53 and others54−66 have used 5′-O-(N-acylsulfamoyl)adenosines (acyl-AMS) and related compounds to inhibit such enzymes by mimicking the cognate, tightly bound acyl-AMP intermediates. Ishida and co-workers first applied this inhibitor design strategy to mechanistically related aminoacyl-tRNA synthetases67 and were inspired by the sulfamoyladenosine class of natural products that includes nucleocidin and ascamycin.68−70 We envisioned that anthranilyl-AMS (1, Table 1) or its sulfamide analogue, anthranilyl-AMSN (2), might be effective inhibitors of PqsA and quinolone biosynthesis. The close structural analogues salicyl-AMS (3),45 salicyl-AMSN (4),59 and benzoyl-AMS (5)59 would provide preliminary structure–activity relationship data. We also envisioned that the corresponding vinyl sulfonamides anthranilyl-AVSN (6) and salicyl-AVSN (7) could inhibit PqsA through covalent binding to the incoming CoA thiol nucleophile during the second half-reaction, forming a mimic of the tetrahedral intermediate.71 This vinyl sulfonamide inhibitor design strategy was previously used by Aldrich and co-workers to target the NRPS salicylate adenylation domain MbtA,71 which is involved in bacterial siderophore biosynthesis, and we have also successfully applied this strategy to an acyl-CoA synthetase MenE, which is involved in bacterial menaquinone biosynthesis,48,49 as well as E1 activating enzymes involved in conjugation of ubiquitin and ubiquitin-like modifier proteins.52,53
Table 1. Inhibition of PqsA by Designed Sulfonyladenosine Inhibitorsa.
| inhibitor | R | X | Kiapp (nM)b |
|---|---|---|---|
| anthranilyl-AMS (1) | NH2 | O | 205 ± 4.0c |
| anthranilyl-AMSN (2) | NH2 | NH | 170 ± 20c |
| salicyl-AMS (3) | OH | O | 88 ± 12 |
| salicyl-AMSN (4) | OH | NH | 109 ± 18 |
| benzoyl-AMS (5) | H | O | 420 ± 69 |
| anthranilyl-AVSN (6) | NH2 | 36300 ± 4300 | |
| salicyl-AVSN (7) | OH | 15400 ± 1700 |
Assays were performed with 60 nM PqsA.
Calculated based on Dixon plots.
Competitive inhibitor with respect to ATP (Ki = 16.5 ± 2.6 nM, calculated based on Morrison equation) and uncompetitive with respect to anthranilate and CoA.
The acyl-AMS sulfamate (1, 3, 5) and acyl-AMSN sulfamide (2, 4) inhibitors were readily synthesized by acylation of a protected sulfamoyladenosine or corresponding sulfamide, respectively, followed by global deprotection using aqueous TFA (see Supporting Information for full details). The sulfamidoadenosine precursor was prepared conveniently by Mitsunobu substitution of the 5′-alcohol of 2′,3′-acetonide-protected adenosine with mono-N-Boc-sulfamide. The acyl-AVSN vinyl sulfonamides (6, 7) were synthesized by Mitsunobu substitution with the corresponding N-Boc-arylvinylsulfonamides directly, followed by deprotection.72
Inhibition of PqsA by Anthranilyl-AMP Analogues
With these inhibitors in hand, we evaluated their ability to inhibit PqsA using a quantitative spectrophotometric assay with recombinant PqsA, detecting formation of anthranilyl-CoA at 365 nm.21 Both anthranilyl-AMS (1) and anthranilyl-AMSN (2) were potent inhibitors of PqsA, with KiATP values of 205 and 170 nM, respectively (Table 1). Further analysis of these “tight-binding inhibitors” using the Morrison equation73,74 indicated that both anthranilyl-AMS (1) and anthranilyl-AMSN (2) are competitive inhibitors with respect to ATP (Ki = 16.5 ± 2.6 nM; 10.5 ± 2.3 nM, respectively) and uncompetitive inhibitors with respect to anthranilate and CoA.
Interestingly, both salicyl-AMS (3) and salicyl-AMSN (4) exhibited potency similar to that of the cognate anthranilyl inhibitors, indicating that an o-phenolic moiety is tolerated by PqsA. This is generally consistent with the previous finding that salicylate, while not accepted as a substrate, does inhibit PqsA.21 Benzoyl-AMS (5) was a somewhat weaker inhibitor, indicating the importance of an ortho substituent on the aromatic ring. This is again consistent with previous biochemical studies indicating that benzoate, while accepted as a PqsA substrate, exhibits a much weaker Km compared to anthranilate (150 μM vs 8 μM).21 In contrast, the corresponding vinyl sulfonamide analogues 6 and 7 were relatively poor inhibitors, at least on the short time scale of this assay. This is consistent with previous studies within this structural superfamily of enzymes, in which we have shown that related vinyl sulfonamides are effective but slow-binding inhibitors of another acyl-CoA synthetase MenE when assayed in the presence of CoA,48,49 while Aldrich et al. have reported that salicyl-AVSN (7) is a weak inhibitor of the salicylate adenylation enzyme MbtA in the absence of the corresponding thiol nucleophile MbtB.71
Inhibition of P. aeruginosa Quinolone Production by Anthranilyl-AMP Analgoues
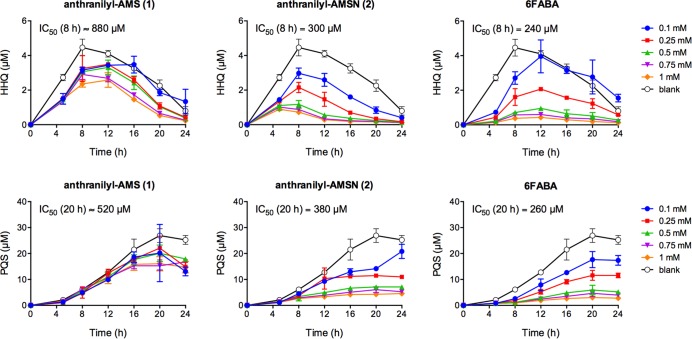
Next, we evaluated the five most potent PqsA inhibitors 1–5 for their ability to inhibit HHQ and PQS quinolone production in P. aeruginosa strain PA14 (Figure 2). HHQ and PQS concentrations were determined by LC-MS/MS quantitation75 relative to deuterated internal standards (see Supporting Information for full details). The vinyl sulfonamide analogues 6 and 7 were excluded due to their lack of biochemical potency. 6-Fluoroanthranilate (6FABA), a substrate analogue reported previously to inhibit quinolone production at millimolar concentrations,42 was used as a positive control. Compounds were initially tested at 1.5 mM concentration, and HHQ and PQS were quantified at 8 and 20 h, respectively. These time points were selected based on an initial time course study of quinolone production in the absence of inhibitors, with HHQ or PQS production peaking at these respective time points. The parent inhibitor anthranilyl-AMS (1) exhibited good inhibition of both HHQ and PQS production under these conditions (67% and 77% inhibition, respectively), whereas the sulfamide analogue anthranilyl-AMSN (2) was more potent (90% and 92% inhibition, respectively), consistent with its increased biochemical potency against PqsA. In contrast, salicyl-AMS (3), salicyl-AMSN (4), and benzoyl-AMS (5) were much less potent inhibitors of quinolone production, despite their similar biochemical potencies against PqsA. This could be due to differences in cell penetration, stability, and/or target specificity. Finally, because inhibitors of the quinolone quorum sensing pathway should not impact bacterial growth, we confirmed that none of the six inhibitors above inhibited growth over 20 h (see Supporting Information Figure S1).
Figure 2.
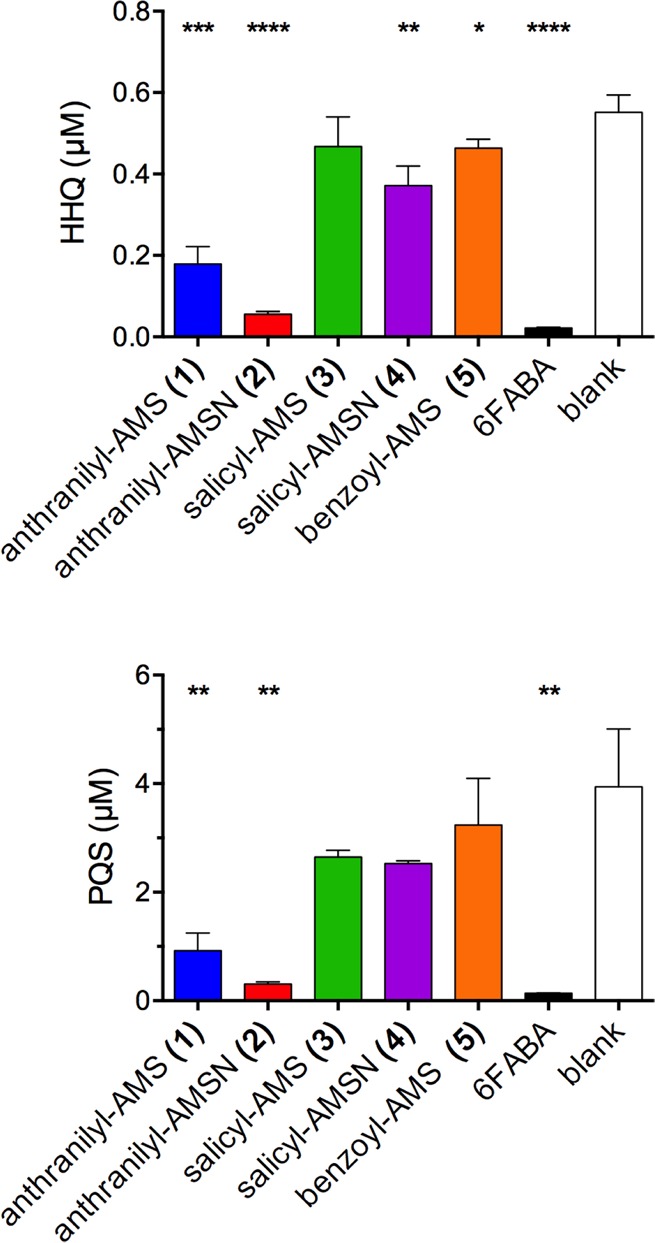
Inhibition of HHQ (8 h) and PQS (20 h) quinolone production in P. aeruginosa strain PA14 (1.5 mM inhibitors). Statistical significance relative to the blank was assessed using a two-tailed unpaired Student t-test with 95% confidence intervals; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Encouraged by the ability of anthranilyl-AMS (1) and anthranilyl-AMSN (2) to inhibit HHQ and PQS quinolone production, we investigated the activity of these two inhibitors and 6FABA in greater detail. In untreated controls, HHQ and PQS production peaked at 8 and 20 h, respectively. Previously reported time course studies have similarly shown that HHQ levels peak at 6–8 h while PQS levels continue to rise.15,76 All three inhibitors were able to decrease quinolone production in a dose- and time-dependent manner (Figure 3). While none of the inhibitors blocked quinolone production completely at any of the concentrations tested, both anthranilyl-AMSN (2) and 6FABA were able to inhibit or delay quinolone production at concentrations as low as 100 μM. Such a perturbation might still be sufficient to impact P. aeruginosa virulence in vivo. Indeed, 6FABA and related haloanthranilate substrate analogues have been shown to increase survival significantly in a mouse burn infection model, despite the fact that they only reduce HHQ levels partially compared to untreated controls in tissue directly underlying the infection site.42 Anthranilyl-AMS (1) exhibited the same trend of inhibition, albeit again with lower potency.
Figure 3.
Dose- and time-dependent inhibition of HHQ (top) and PQS (bottom) quinolone production in P. aeruginosa strain PA14 over 24 h.
Effect of Anthranilyl-AMP Analogues on P. aeruginosa Pyocyanin Production
We next investigated the ability of these PqsA inhibitors to block production of the virulence factor pyocyanin. Although both anthranilyl-AMS (1) and anthranilyl-AMSN (2) were able to reduce HHQ and PQS production at 1 mM concentration (1, 47 and 40% inhibition, respectively; 2, 84% and 84% inhibition, respectively; Figure 3), neither inhibited pyocyanin production at this concentration (see Supporting Information Figure S2). Of note, MvfR antagonists37 and PqsD inhibitors25,38 that similarly inhibit quinolone production but not pyocyanin production have been reported previously. Haloanthranilate substrate analogues have been reported to inhibit both quinolone and pyocyanin production but may impact multiple targets in this complex signaling network.42 This is indicative of the complex relationship between HHQ and PQS quinolone biosynthesis, induction of the MvfR regulon, and production of pyocyanin.
Compound Accumulation of Anthranilyl-AMP Analogues in P. aeruginosa
It is well-known that P. aeruginosa has high intrinsic resistance to small-molecule antibiotics due to limited membrane permeability and high efflux.3,4 We hypothesized that this may be the basis for the difference between the potent biochemical activity of our PqsA inhibitors and their modest cellular potencies in P. aeruginosa. Thus, we carried out compound accumulation studies with anthranilyl-AMS (1), anthranilyl-AMSN (2), and 6FABA in P. aeruginosa PA14 using an LC-MS/MS quantitation approach (see Figure 4).77 Cells were grown in LB to early log phase (OD600 ≈ 0.5), treated with 1000 μM compound for 30 min, then centrifuged and rapidly washed four times with cold PBS to remove surface-associated compound. The cells were lysed by multiple freeze–thaw cycles, centrifuged, and precipitated with methanol containing an internal standard (benzoyl-AMS, 5). Compound concentration in the lysate was quantitated by LC-MS/MS relative to the internal standard and the intracellular concentration was calculated based on CFU determination of the culture just prior to washing. Anthranilyl-AMS (1), anthranilyl-AMSN (2), and 6FABA accumulated to 20–30 μM intracellular concentration under these conditions, corresponding to 2–3% penetration relative to the 1000 μM extracellular concentration applied. These results suggest that limited penetration of anthranilyl-AMS (1) and anthranilyl-AMSN (2) into P. aeruginosa contributes to the differences between their biochemical and cellular potencies observed above, although other factors such as metabolic stability and/or off-target binding must also be considered. These integrated data provide a basis to correlate biochemical inhibition, bacterial compound accumulation, and cellular activity and indicate that membrane penetration and/or efflux evasion are keys to improving the cellular activity of these rationally designed PqsA inhibitors.
Figure 4.
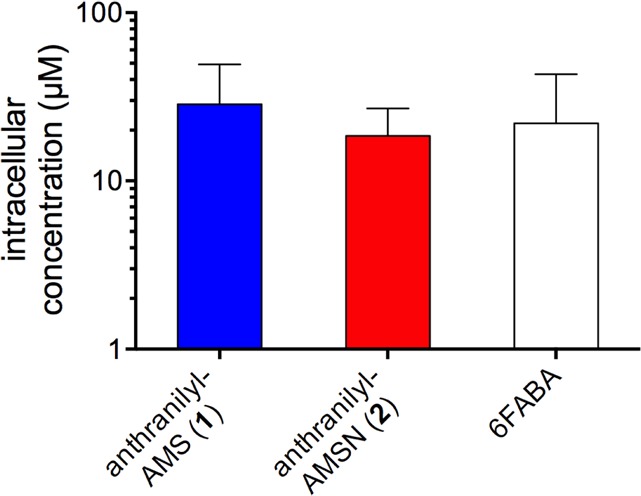
Compound accumulation in P. aeruginosa strain PA14 after incubation with 1000 μM extracellular concentration for 30 min.
Conclusions
The P. aeruginosa quinolone signaling system is an attractive target for the development of novel therapeutics that block virulence factor production. We have developed a series of rationally designed inhibitors of the anthranilyl-CoA synthetase PqsA, which catalyzes a key step in P. aeruginosa quinolone biosynthesis. Anthranilyl-AMS (1) and anthranilyl-AMSN (2) were identified as potent biochemical inhibitors of PqsA having moderate inhibitory activity against HHQ and PQS quinolone production in P. aeruginosa. Compound accumulation studies suggest that the limited cell penetration of these inhibitors may contribute to this difference in biochemical versus cellular activities. This work provides direct pharmacological validation of PqsA as a promising target to inhibit HHQ and PQS quinolone biosynthesis in P. aeruginosa. Future efforts will focus on developing analogues with improved cell penetrance or combinations with efflux pump inhibitors or membrane-permeablizing antibiotics to provide more potent cellular activity. These inhibitors will then be useful probes for investigating the interplay between quinolone signaling and virulence factor production in P. aeruginosa, including quantitative assessment of the level and timing of virulence factor inhibition required to impact in vivo virulence, as well as evaluation in conjunction with other antibiotics, particularly those targeting the interconnected LasR- and RhlR-regulated quorum sensing systems,78 with the long-term goal of developing novel antivirulence strategies to treat P. aeruginosa infections.
Acknowledgments
We thank G. Chiosis and T. Taldone (MSKCC) for providing access to LC-MS/MS instrumentation and G. Sukenick, R. Wang, H. Liu, H. Fang, and S. Rusli (MSKCC) for expert mass spectrometry support. Financial support from the National Institutes of Health (R21/R33 AI098802 to D.S.T. and L.G.R., R01 AI068038 to D.S.T., R21/R33 AI105902 to L.G.R., R01 AI076272 to E.C.P. and J.P.C., Cancer Center Support Grant P30 CA008748 to C. B. Thompson) and the Lucille Castori Center for Microbes, Inflammation, and Cancer (postdoctoral fellowship to I. S.) is gratefully acknowledged.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.6b00575.
Complete experimental procedures and analytical data for all new compounds (PDF)
The authors declare the following competing financial interest(s): D.S.T. is a coinventor of issued US patents and patent applications on sulfonyladenosines as antibacterial agents and has intellectual property interests in these compounds. L.G.R. is the scientific founder and a scientific advisory board member of Spero Therapeutics LLC. No funding from Spero Therapeutics was received.
Supplementary Material
References
- Page M. G. P.; Heim J. (2009) Prospects for the next anti-Pseudomonas drug. Curr. Opin. Pharmacol. 9, 558–565. 10.1016/j.coph.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Gomez M. I.; Prince A. (2007) Opportunistic infections in lung disease. Curr. Opin. Pharmacol. 7, 244–251. 10.1016/j.coph.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Bonomo R. A.; Szabo D. (2006) Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43, S49–S54. 10.1086/504477. [DOI] [PubMed] [Google Scholar]
- Zavascki A. P.; Carvalhaes C. G.; Picao R. C.; Gales A. C. (2010) Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: Resistance mechanisms and implications for therapy. Expert Rev. Anti-Infect. Ther. 8, 71–93. 10.1586/eri.09.108. [DOI] [PubMed] [Google Scholar]
- O’Connell K. M. G.; Hodgkinson J. T.; Sore H. F.; Welch M.; Salmond G. P. C.; Spring D. R. (2013) Combating multidrug-resistant bacteria: Current strategies for the discovery of novel antibacterials. Angew. Chem., Int. Ed. 52, 10706–10733. 10.1002/anie.201209979. [DOI] [PubMed] [Google Scholar]
- Galloway W. R. J. D.; Hodgkinson J. T.; Bowden S.; Welch M.; Spring D. R. (2012) Applications of small molecule activators and inhibitors of quorum sensing in Gram-negative bacteria. Trends Microbiol. 20, 449–458. 10.1016/j.tim.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Heras B.; Scanlon M. J.; Martin J. L. (2015) Targeting virulence not viability in the search for future antibacterials. Br. J. Clin. Pharmacol. 79, 208–215. 10.1111/bcp.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSarre B.; Federle M. J. (2013) Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 77, 73–111. 10.1128/MMBR.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal Jimenez P.; Koch G.; Thompson J. A.; Xavier K. B.; Cool R. H.; Quax W. J. (2012) The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 76, 46–65. 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P.; Camara M. (2009) Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 12, 182–191. 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Pesci E. C.; Milbank J. B. J.; Pearson J. P.; McKnight S.; Kende A. S.; Greenberg E. P.; Iglewski B. H. (1999) Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96, 11229–11234. 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G.; Deziel E.; He J.; Lepine F.; Lesic B.; Castonguay M.-H.; Milot S.; Tampakaki A. P.; Stachel S. E.; Rahme L. G. (2006) MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol. Microbiol. 62, 1689–1699. 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- Calfee M. W.; Coleman J. P.; Pesci E. C. (2001) Interference with Pseudomonas quinolone signal synthesis inhibits virulence factor expression by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 98, 11633–11637. 10.1073/pnas.201328498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H.; Krishnan G.; Goumnerov B.; Tsongalis J.; Tompkins R.; Rahme L. G. (2001) A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. U. S. A. 98, 14613–14618. 10.1073/pnas.251465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deziel E.; Lepine F.; Milot S.; He J.; Mindrinos M. N.; Tompkins R. G.; Rahme L. G. (2004) Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. U. S. A. 101, 1339–1344. 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deziel E.; Gopalan S.; Tampakaki A. P.; Lepine F.; Padfield K. E.; Saucier M.; Xiao G.; Rahme L. G. (2005) The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: Multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol. Microbiol. 55, 998–1014. 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- McGrath S.; Wade D. S.; Pesci E. C. (2004) Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol. Lett. 230, 27–34. 10.1016/S0378-1097(03)00849-8. [DOI] [PubMed] [Google Scholar]
- McKnight S. L.; Iglewski B. H.; Pesci E. C. (2000) The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182, 2702–2708. 10.1128/JB.182.10.2702-2708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulcey C. E.; Dekimpe V.; Fauvelle D.-A.; Milot S.; Groleau M.-C.; Doucet N.; Rahme L. G.; Lepine F.; Deziel E. (2013) The end of an old hypothesis: The Pseudomonas signaling molecules 4-hydroxy-2-alkylquinolines derive from fatty acids, not 3-ketofatty acids. Chem. Biol. 20, 1481–1491. 10.1016/j.chembiol.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubern J.-F.; Diggle S. P. (2008) Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol. BioSyst. 4, 882–888. 10.1039/b803796p. [DOI] [PubMed] [Google Scholar]
- Coleman J. P.; Hudson L. L.; McKnight S. L.; Farrow J. M. III; Calfee M. W.; Lindsey C. A.; Pesci E. C. (2008) Pseudomonas aeruginosa PqsA is an anthranilate-coenzyme A ligase. J. Bacteriol. 190, 1247–1255. 10.1128/JB.01140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistorius D.; Ullrich A.; Lucas S.; Hartmann R. W.; Kazmaier U.; Mueller R. (2011) Biosynthesis of 2-alkyl-4(1H)-quinolones in Pseudomonas aeruginosa: Potential for therapeutic interference with pathogenicity. ChemBioChem 12, 850–853. 10.1002/cbic.201100014. [DOI] [PubMed] [Google Scholar]
- Weidel E.; de Jong J. C.; Brengel C.; Storz M. P.; Braunshausen A.; Negri M.; Plaza A.; Steinbach A.; Mueller R.; Hartmann R. W. (2013) Structure optimization of 2-benzamidobenzoic acids as PqsD inhibitors for Pseudomonas aeruginosa Infections and elucidation of binding mode by SPR, STD NMR, and molecular docking. J. Med. Chem. 56, 6146–6155. 10.1021/jm4006302. [DOI] [PubMed] [Google Scholar]
- Hinsberger S.; de Jong J. C.; Groh M.; Haupenthal J.; Hartmann R. W. (2014) Benzamidobenzoic acids as potent PqsD inhibitors for the treatment of Pseudomonas aeruginosa infections. Eur. J. Med. Chem. 76, 343–351. 10.1016/j.ejmech.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Storz M. P.; Maurer C. K.; Zimmer C.; Wagner N.; Brengel C.; de Jong J. C.; Lucas S.; Muesken M.; Haeussler S.; Steinbach A.; Hartmann R. W. (2012) Validation of PqsD as an anti-biofilm target in Pseudomonas aeruginosa by development of small-molecule Inhibitors. J. Am. Chem. Soc. 134, 16143–16146. 10.1021/ja3072397. [DOI] [PubMed] [Google Scholar]
- Storz M. P.; Brengel C.; Weidel E.; Hoffmann M.; Hollemeyer K.; Steinbach A.; Mueller R.; Empting M.; Hartmann R. W. (2013) Biochemical and biophysical analysis of a chiral PqsD inhibitor revealing tight-binding behavior and enantiomers with contrary thermodynamic signatures. ACS Chem. Biol. 8, 2794–2801. 10.1021/cb400530d. [DOI] [PubMed] [Google Scholar]
- Storz M. P.; Allegretta G.; Kirsch B.; Empting M.; Hartmann R. W. (2014) From in vitro to in cellulo: Structure–activity relationship of (2-nitrophenyl) methanol derivatives as inhibitors of PqsD in Pseudomonas aeruginosa. Org. Biomol. Chem. 12, 6094–6104. 10.1039/C4OB00707G. [DOI] [PubMed] [Google Scholar]
- Sahner J. H.; Brengel C.; Storz M. P.; Groh M.; Plaza A.; Mueller R.; Hartmann R. W. (2013) Combining in silico and biophysical methods for the development of Pseudomonas aeruginosa quorum sensing Inhibitors: An alternative approach for structure-based drug design. J. Med. Chem. 56, 8656–8664. 10.1021/jm401102e. [DOI] [PubMed] [Google Scholar]
- Sahner J. H.; Empting M.; Kamal A.; Weidel E.; Groh M.; Boerger C.; Hartmann R. W. (2015) Exploring the chemical space of ureidothiophene-2-carboxylic acids as inhibitors of the quorum sensing enzyme PqsD from Pseudomonas aeruginosa. Eur. J. Med. Chem. 96, 14–21. 10.1016/j.ejmech.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Zender M.; Witzgall F.; Drees S. L.; Weidel E.; Maurer C. K.; Fetzner S.; Blankenfeldt W.; Empting M.; Hartmann R. W. (2016) Dissecting the multiple roles of PqsE in Pseudomonas aeruginosa virulence by discovery of small tool compounds. ACS Chem. Biol. 11, 1755–1763. 10.1021/acschembio.6b00156. [DOI] [PubMed] [Google Scholar]
- Lu C.; Kirsch B.; Zimmer C.; de Jong J. C.; Henn C.; Maurer C. K.; Muesken M.; Haeussler S.; Steinbach A.; Hartmann R. W. (2012) Discovery of antagonists of PqsR, a key player in 2-alkyl-4-quinolone-dependent quorum sensing in Pseudomonas aeruginosa. Chem. Biol. 19, 381–390. 10.1016/j.chembiol.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Klein T.; Henn C.; de Jong J. C.; Zimmer C.; Kirsch B.; Maurer C. K.; Pistorius D.; Mueller R.; Steinbach A.; Hartmann R. W. (2012) Identification of small-molecule antagonists of the Pseudomonas aeruginosa transcriptional regulator PqsR: Biophysically guided hit discovery and optimization. ACS Chem. Biol. 7, 1496–1501. 10.1021/cb300208g. [DOI] [PubMed] [Google Scholar]
- Lu C.; Maurer C. K.; Kirsch B.; Steinbach A.; Hartmann R. W. (2014) Overcoming the unexpected functional inversion of a PqsR antagonist in Pseudomonas aeruginosa: An in vivo potent antivirulence agent targeting pqs quorum sensing. Angew. Chem., Int. Ed. 53, 1109–1112. 10.1002/anie.201307547. [DOI] [PubMed] [Google Scholar]
- Lu C.; Kirsch B.; Maurer C. K.; de Jong J. C.; Braunshausen A.; Steinbach A.; Hartmann R. W. (2014) Optimization of anti-virulence PqsR antagonists regarding aqueous solubility and biological properties resulting in new insights in structure–activity relationships. Eur. J. Med. Chem. 79, 173–183. 10.1016/j.ejmech.2014.04.016. [DOI] [PubMed] [Google Scholar]
- Zender M.; Klein T.; Henn C.; Kirsch B.; Maurer C. K.; Kail D.; Ritter C.; Dolezal O.; Steinbach A.; Hartmann R. W. (2013) Discovery and biophysical characterization of 2-amino-oxadiazoles as novel antagonists of PqsR, an important regulator of Pseudomonas aeruginosa virulence. J. Med. Chem. 56, 6761–6774. 10.1021/jm400830r. [DOI] [PubMed] [Google Scholar]
- Ilangovan A.; Fletcher M.; Rampioni G.; Pustelny C.; Rumbaugh K.; Heeb S.; Camara M.; Truman A.; Chhabra S. R.; Emsley J.; Williams P. (2013) Structural basis for native agonist and synthetic inhibitor recognition by the Pseudomonas aeruginosa quorum sensing regulator PqsR (MvfR). PLoS Pathog. 9, e1003508. 10.1371/journal.ppat.1003508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey M.; Lepine F.; Maura D.; Bandyopadhaya A.; Lesic B.; He J.; Kitao T.; Righi V.; Milot S.; Tzika A.; Rahme L. (2014) Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog. 10, e1004321. 10.1371/journal.ppat.1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann A.; de Mello Martins A. G. G.; Brengel C.; Empting M.; Hartmann R. W. (2016) Application of dual inhibition concept within looped autoregulatory systems toward antivirulence agents against Pseudomonas aeruginosa infections. ACS Chem. Biol. 11, 1279–1286. 10.1021/acschembio.6b00117. [DOI] [PubMed] [Google Scholar]
- Kasper S. H.; Bonocora R. P.; Wade J. T.; Musah R. A.; Cady N. C. (2016) Chemical inhibition of kynureninase reduces Pseudomonas aeruginosa quorum sensing and virulence factor expression. ACS Chem. Biol. 11, 1106–1117. 10.1021/acschembio.5b01082. [DOI] [PubMed] [Google Scholar]
- Kim K.; Kim Y. U.; Koh B. H.; Hwang S. S.; Kim S.-H.; Lepine F.; Cho Y.-H.; Lee G. R. (2010) HHQ and PQS, two Pseudomonas aeruginosa quorum-sensing molecules, down-regulate the innate immune responses through the nuclear factor-κB pathway. Immunology 129, 578–588. 10.1111/j.1365-2567.2009.03160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müsken M.; Di Fiore S.; Dötsch A.; Fischer R.; Häussler S. (2010) Genetic determinants of Pseudomonas aeruginosa biofilm establishment. Microbiology 156, 431–441. 10.1099/mic.0.033290-0. [DOI] [PubMed] [Google Scholar]
- Lesic B.; Lepine F.; Deziel E.; Zhang J.; Zhang Q.; Padfield K.; Castonguay M.-H.; Milot S.; Stachel S.; Tzika A. A.; Tompkins R. G.; Rahme L. G. (2007) Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog. 3, e126. 10.1371/journal.ppat.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor U.; Bielecki P.; Loessner H.; Rohde M.; Wolf K.; Westphal K.; Weiss S.; Haeussler S. (2012) Biofilm formation by Pseudomonas aeruginosa in solid murine tumors - A novel model system. Microbes Infect. 14, 951–958. 10.1016/j.micinf.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Gulick A. M. (2009) Conformational dynamics in the acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811–827. 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreras J. A.; Ryu J.-S.; Di Lello F.; Tan D. S.; Quadri L. E. N. (2005) Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis. Nat. Chem. Biol. 1, 29–32. 10.1038/nchembio706. [DOI] [PubMed] [Google Scholar]
- Lun S.; Guo H.; Adamson J.; Cisar J. S.; Davis T. D.; Chavadi S. S.; Warren J. D.; Quadri L. E. N.; Tan D. S.; Bishai W. R. (2013) Pharmacokinetic and in vivo efficacy studies of the mycobactin biosynthesis inhibitor salicyl-AMS in mice. Antimicrob. Agents Chemother. 57, 5138–5140. 10.1128/AAC.00918-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisar J. S.; Ferreras J. A.; Soni R. K.; Quadri L. E. N.; Tan D. S. (2007) Exploiting ligand conformation in the selective inhibition of non-ribosomal peptide synthetase amino acid adenylation with designed macrocyclic small molecules. J. Am. Chem. Soc. 129, 7752–7753. 10.1021/ja0721521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.; Zhang H.; Tonge P. J.; Tan D. S. (2008) Mechanism-based inhibitors of MenE, an acyl-CoA synthetase involved in bacterial menaquinone biosynthesis. Bioorg. Med. Chem. Lett. 18, 5963–5966. 10.1016/j.bmcl.2008.07.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.; Zhou R.; Sharma I.; Li X.; Kumar G.; Swaminathan S.; Tonge P. J.; Tan D. S. (2012) Stable analogues of OSB-AMP: Potent inhibitors of MenE, the o-succinylbenzoate-CoA synthetase from bacterial menaquinone biosynthesis. ChemBioChem 13, 129–136. 10.1002/cbic.201100585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarlo J. S.; Evans C. E.; Sharma I.; Lavaud L. J.; Ngo S. C.; Shek R.; Rajashankar K. R.; French J. B.; Tan D. S.; Tonge P. J. (2015) Mechanism of MenE inhibition by acyl-adenylate analogues and discovery of novel antibacterial agents. Biochemistry 54, 6514–6524. 10.1021/acs.biochem.5b00966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreras J. A.; Stirrett K. L.; Lu X.; Ryu J.-S.; Soll C. E.; Tan D. S.; Quadri L. E. N. (2008) Mycobacterial phenolic glycolipid virulence factor biosynthesis: Mechanism and small-molecule inhibition of polyketide chain initiation. Chem. Biol. 15, 51–61. 10.1016/j.chembiol.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.; Olsen S. K.; Capili A. D.; Cisar J. S.; Lima C. D.; Tan D. S. (2010) Designed semisynthetic protein inhibitors of Ub/Ubl E1 activating enzymes. J. Am. Chem. Soc. 132, 1748–1749. 10.1021/ja9088549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen S. K.; Capili A. D.; Lu X.; Tan D. S.; Lima C. D. (2010) Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature 463, 906–912. 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finking R.; Neumueller A.; Solsbacher J.; Konz D.; Kretzschmar G.; Schweitzer M.; Krumm T.; Marahiel M. A. (2003) Aminoacyl adenylate substrate analogues for the inhibition of adenylation domains of nonribosomal peptide synthetases. ChemBioChem 4, 903–906. 10.1002/cbic.200300666. [DOI] [PubMed] [Google Scholar]
- Koroniak L.; Ciustea M.; Gutierrez J. A.; Richards N. G. J. (2003) Synthesis and characterization of an N-acylsulfonamide inhibitor of human asparagine synthetase. Org. Lett. 5, 2033–2036. 10.1021/ol034212n. [DOI] [PubMed] [Google Scholar]
- Brown P. H.; Beckett D. (2005) Use of binding enthalpy to drive an allosteric transition. Biochemistry 44, 3112–3121. 10.1021/bi047792k. [DOI] [PubMed] [Google Scholar]
- Branchini B. R.; Murtiashaw M. H.; Carmody J. N.; Mygatt E. E.; Southworth T. L. (2005) Synthesis of an N-acyl sulfamate analog of luciferyl-AMP: A stable and potent inhibitor of firefly luciferase. Bioorg. Med. Chem. Lett. 15, 3860–3864. 10.1016/j.bmcl.2005.05.115. [DOI] [PubMed] [Google Scholar]
- May J. J.; Finking R.; Wiegeshoff F.; Weber T. T.; Bandur N.; Koert U.; Marahiel M. A. (2005) Inhibition of the D-alanine:D-alanyl carrier protein ligase from Bacillus subtilis increases the bacterium’s susceptibility to antibiotics that target the cell wall. FEBS J. 272, 2993–3003. 10.1111/j.1742-4658.2005.04700.x. [DOI] [PubMed] [Google Scholar]
- Somu R. V.; Boshoff H.; Qiao C.; Bennett E. M.; Barry C. E. III; Aldrich C. C. (2006) Rationally designed nucleoside antibiotics that inhibit siderophore biosynthesis of Mycobacterium tuberculosis. J. Med. Chem. 49, 31–34. 10.1021/jm051060o. [DOI] [PubMed] [Google Scholar]
- Miethke M.; Bisseret P.; Beckering C. L.; Vignard D.; Eustache J.; Marahiel M. A. (2006) Inhibition of aryl acid adenylation domains involved in bacterial siderophore synthesis. FEBS J. 273, 409–419. 10.1111/j.1742-4658.2005.05077.x. [DOI] [PubMed] [Google Scholar]
- Pfleger B. F.; Lee J. Y.; Somu R. V.; Aldrich C. C.; Hanna P. C.; Sherman D. H. (2007) Characterization and analysis of early enzymes for petrobactin biosynthesis in Bacillus anthracis. Biochemistry 46, 4147–4157. 10.1021/bi6023995. [DOI] [PubMed] [Google Scholar]
- Tian Y.; Suk D.-H.; Cai F.; Crich D.; Mesecar A. D. (2008) Bacillus anthracis o-succinylbenzoyl-CoA synthetase: Reaction kinetics and a novel inhibitor mimicking its reaction intermediate. Biochemistry 47, 12434–12447. 10.1021/bi801311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciulli A.; Scott D. E.; Ando M.; Reyes F.; Saldanha S. A.; Tuck K. L.; Chirgadze D. Y.; Blundell T. L.; Abell C. (2008) Inhibition of Mycobacterium tuberculosis pantothenate synthetase by analogues of the reaction intermediate. ChemBioChem 9, 2606–2611. 10.1002/cbic.200800437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P.; Goyal A.; Natarajan V. T.; Rajakumara E.; Verma P.; Gupta R.; Yousuf M.; Trivedi O. A.; Mohanty D.; Tyagi A.; Sankaranarayanan R.; Gokhale R. S. (2009) Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis. Nat. Chem. Biol. 5, 166–173. 10.1038/nchembio.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake E. J.; Duckworth B. P.; Neres J.; Aldrich C. C.; Gulick A. M. (2010) Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis. Biochemistry 49, 9292–9305. 10.1021/bi101226n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora A. L.; Wilson D. J.; Aldrich C. C.; Blanchard J. S. (2010) Kinetic and inhibition studies of dihydroxybenzoate-AMP ligase from Escherichia coli. Biochemistry 49, 3648–3657. 10.1021/bi100350c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H.; Shoku Y.; Hayashi N.; Mitsunaga J.; In Y.; Doi M.; Inoue M.; Ishida T. (1991) X-ray crystallographic conformational study of 5′-O-[N-(L-alanyl)-sulfamoyl]adenosine, a substrate analog for alanyl-tRNA synthetase. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 1080, 126–134. 10.1016/0167-4838(91)90138-P. [DOI] [PubMed] [Google Scholar]
- Waller C. W.; Patrick J. B.; Fulmor W.; Meyer W. E. (1957) The structure of nucleocidin. I. J. Am. Chem. Soc. 79, 1011–1012. 10.1021/ja01561a076. [DOI] [Google Scholar]
- Morton G. O.; Lancaster J. E.; Van Lear G. E.; Fulmor W.; Meyer W. E. (1969) Structure of nucleocidin. III. Revised structure. J. Am. Chem. Soc. 91, 1535–1537. 10.1021/ja01034a046. [DOI] [PubMed] [Google Scholar]
- Isono K.; Uramoto M.; Kusakabe H.; Miyata N.; Koyama T.; Ubukata M.; Sethi S. K.; McCloskey J. A. (1984) Ascamycin and dealanylascamycin, nucleoside antibiotics from Streptomyces sp. J. Antibiot. 37, 670–672. 10.7164/antibiotics.37.670. [DOI] [PubMed] [Google Scholar]
- Qiao C.; Wilson D. J.; Bennett E. M.; Aldrich C. C. (2007) A mechanism-based aryl carrier protein/thiolation domain affinity probe. J. Am. Chem. Soc. 129, 6350–6351. 10.1021/ja069201e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundlov J. A.; Shi C.; Wilson D. J.; Aldrich C. C.; Gulick A. M. (2012) Structural and functional investigation of the intermolecular interaction between NRPS adenylation and carrier protein domains. Chem. Biol. 19, 188–198. 10.1016/j.chembiol.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison J. F. (1969) Kinetics of the reversible inhibition of enzyme-catalyzed reactions by tight-binding inhibitors. Biochim. Biophys. Acta, Enzymol. 185, 269–286. 10.1016/0005-2744(69)90420-3. [DOI] [PubMed] [Google Scholar]
- Williams J. W.; Morrison J. F. (1979) The kinetics of reversible tight-binding inhibition. Methods Enzymol. 63, 437–467. 10.1016/0076-6879(79)63019-7. [DOI] [PubMed] [Google Scholar]
- Lepine F.; Deziel E.; Milot S.; Rahme L. G. (2003) A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim. Biophys. Acta, Gen. Subj. 1622, 36–41. 10.1016/S0304-4165(03)00103-X. [DOI] [PubMed] [Google Scholar]
- Kesarwani M.; Hazan R.; He J.; Que Y.; Apidianakis Y.; Lesic B.; Xiao G.; Dekimpe V.; Milot S.; Deziel E.; Lepine F.; Rahme L. G. (2011) A quorum sensing regulated small volatile molecule reduces acute virulence and promotes chronic infection phenotypes. PLoS Pathog. 7, e1002192. 10.1371/journal.ppat.1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. D.; Gerry C. J.; Tan D. S. (2014) General platform for systematic quantitative evaluation of small-molecule permeability in bacteria. ACS Chem. Biol. 9, 2535–2544. 10.1021/cb5003015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh M. A.; Blackwell H. E. (2016) Chemical genetics reveals environment-specific roles for quorum sensing circuits in Pseudomonas aeruginosa. Cell Chem. Biol. 23, 361–369. 10.1016/j.chembiol.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.