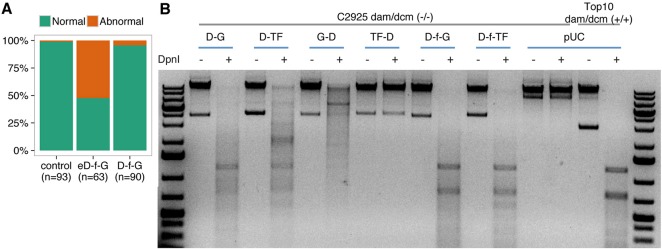
Fig. 1.
Improving Dam-fusion proteins. (A) DamL122A displays low toxicity in medaka embryos compared with the unmodified protein. Medaka zygotes were injected with mRNA coding for the E. coli Dam (eD-f-G) or DamL122A fused to GFP via flexylinker (D-f-G) (see below). Embryos were scored for abnormalities at embryonic stage 25. (B) Agarose gel of isolated bacterial gDNA samples undigested (−) or digested (+) with DpnI. Dam activity depends on the flexilinker, and the type and orientation of the fused proteins. Bacterial gDNA isolated from a strain deficient in the dam/dcm systems is resistant to DpnI digestion. This condition can be reversed in transformed bacteria only when the fusion protein generates a functional Dam. Whereas DNA from bacteria transformed with constructs coding for fusions Dam-GFP (D-G) or Dam-TF (D-TF) (OtpA from zebrafish) can be digested by DpnI, DNA from GFP-Dam (G-D) and TF-Dam (TF-D) bacteria is resistant to DpnI digestion. In addition, the use of flexylinker between Dam and the fusion protein (D-f-GFP and D-f-TF) generates a DpnI digestion pattern similar to that of bacteria with a functional dam/dcm system (Top10 cells).