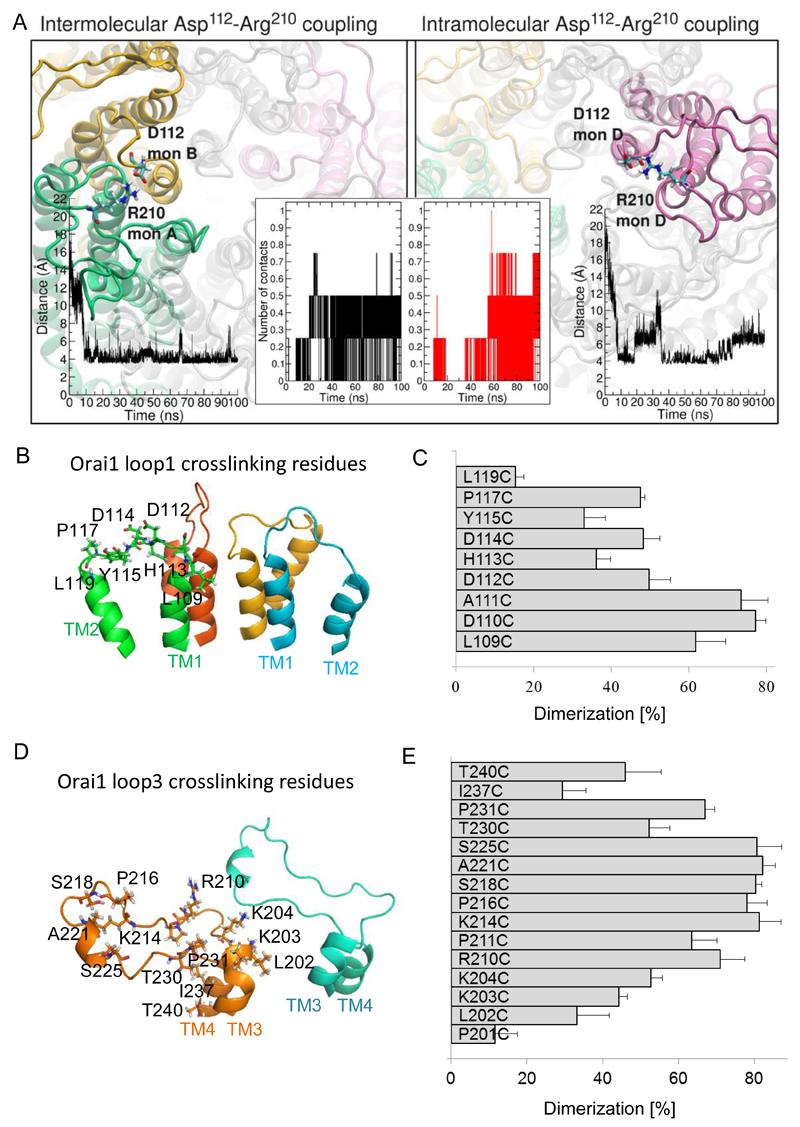
Figure 5. Loop3 residues form electrostatic interactions with CAR in Orai1.
(A) Representative snapshot of the intermolecular (left panel) and intramolecular (right panel) electrostatic interaction of Arg210 with Asp112 predicted by molecular dynamics simulation. Potential Asp112-Arg210 contacts were counted based on a distance ≤ 4.5 Å between the carboxyl carbon of loop1 aspartic acid and the guanidino carbon of loop3 arginine as shown in the time-courses (left inset graph for intermolecular interaction, and right inset graph for intramolecular interaction). The time-dependent number of intramolecular and intermolecular contacts between Asp112 in loop1 and Arg210 in loop3 for a single Orai1 channel within 100 ns is shown in the central inset. (B) Selected single cysteine mutants in Orai1 loop1 (L109C, D110C, A111C, D112C, H113C, D114C, P117C, L110C) are shown as sticks in a 3D homology model of Orai1. (C) Percentage of dimerization for cysteine mutants upon CuP treatment were calculated. Data are shown as the average + SEM (n = 4 -8). (D) Structural modelling of the loop3 domain in Orai1 yielded a random coil region, highlighting the residues (P201, L202, K203, K204, R210, P211, K214, P216, S218, A221, S225, T230, P231, I237, T240), which were individually engineered to cysteines. (E) Percentage of dimerization for cysteine mutants upon CuP treatment were calculated. Data are shown as the average + SEM (n = 4 – 8).