Fig 3. Phenotypic and genotypic characterization of K. pneumoniae strains from medical centre in India.
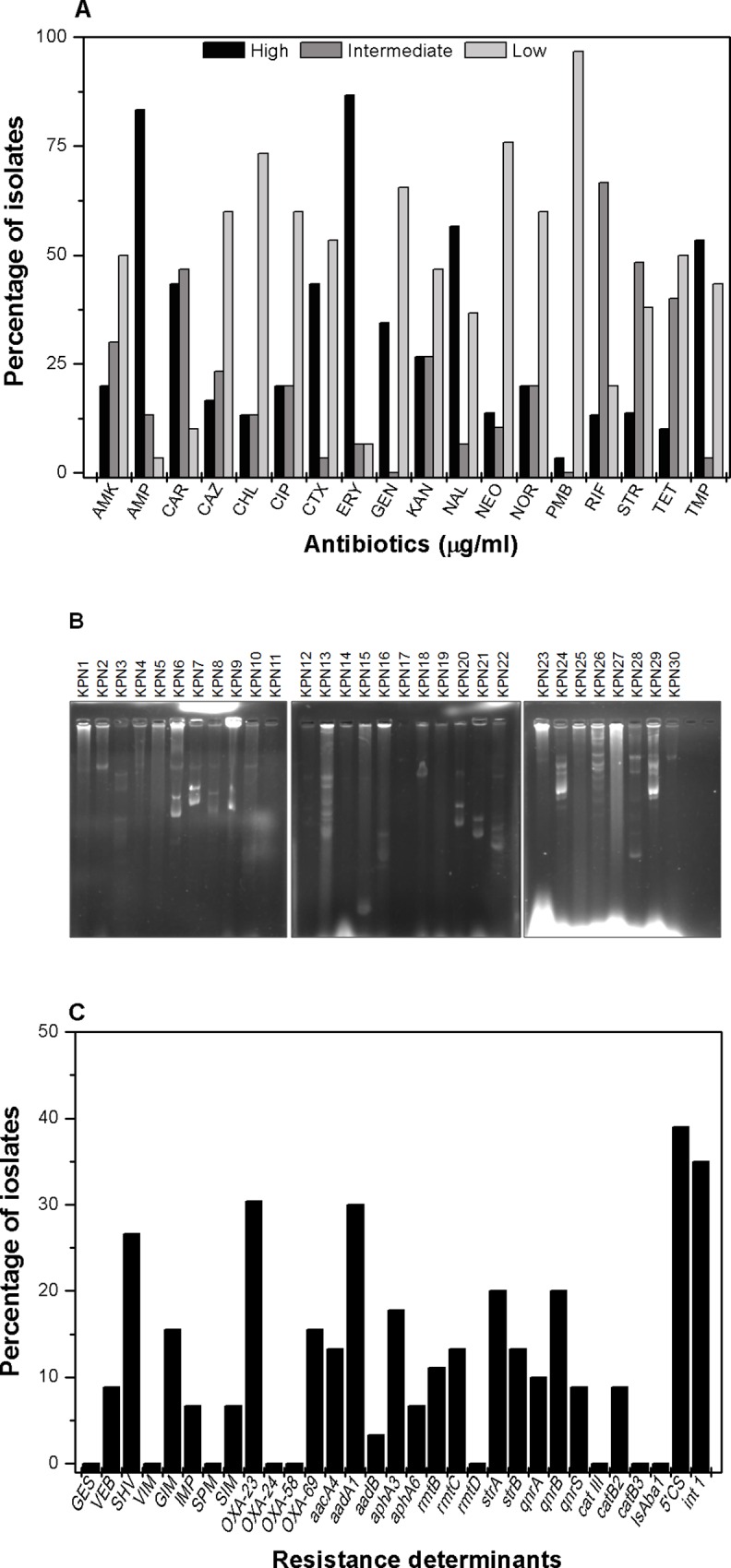
(A) Graphical presentation of percentage of isolates with different level of resistance with different antibiotics. In this study we grouped the isolates in three different resistance level for each antibiotic viz. strains with high MIC values (256 μg/ml to >1024 μg/ml) are grouped as highly resistant, intermediate MIC value (16 μg/ml to 64 μg/ml) as intermediately resistant and low MIC value (4 μg/ml or less) as sensitive. For this study, we used the following antibiotics-AMP: ampicillin, CAR: carbenicillin, CTR: ceftriaxone, CAZ: ceftazidime, AMK: amikacin, GEN: gentamicin, KAN: kanamycin NEO: neomycin, STR: streptomycin CIP: ciprofloxacin, NAL: nalidixic acid, NOR: norfloxacin, CHL: chloramphenicol, ERY: erythromycin, PMB: polymyxin B, RIF: rifampicin, TET: tetracycline, TMP: trimethoprim. The bar graph represents the mean of three independent experiments. (B) DNA gel picture of K. pneumoniae plasmids isolated by alkaline lysis method from respective strains showed multiple plasmids of different size. The MDR strains carried multiple plasmids which indicated plasmids might play role in drug resistance. (C) The prevalence to different resistance determinants in the K. pneumoniae isolates are shown here. In this study, we found high prevalence of β-lactamases (blaGES, blaVEB, blaSHV, blaVIM, blaGIM, blaIMP, bla SPM, blaSIM, blaoxa 23, blaoxa 24, blaoxa 24, blaoxa 58, blaOXA69); aminoglycosidases(aacA4, aadA1, aadB, aphA3, aphA6, rmtB, rmtC, rmtD, strA, strB) and quinolone resistance genes qnrA, qnrB, qnrS, including cat III, catB2, catB3, IsAba1, 5' CS of Class 1 integrons.
