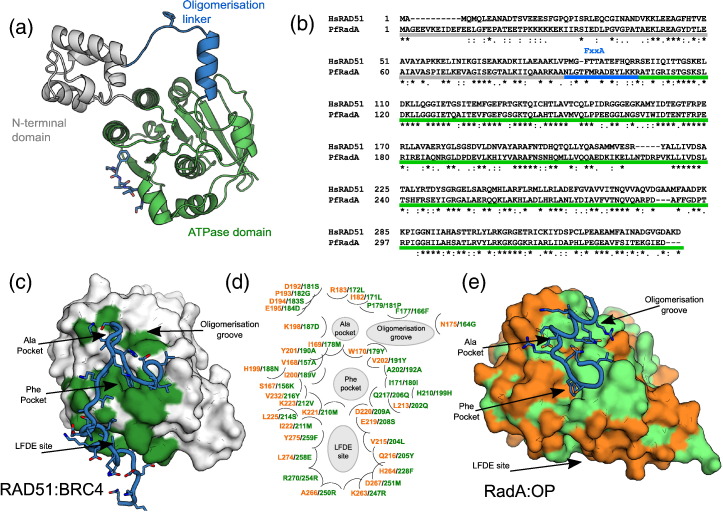
Fig. 1.
Comparative analysis of HsRAD51 and PfRadA. (a) Domain structure of RAD51 with N-terminal domain in grey, FxxA containing linker in blue, and ATPase domain in green. (b) Alignment of human RAD51 and P. furiosus RadA, with different domains highlighted in the same colours as the structure in panel (a). Asterisks indicate identical residues between the two proteins. (c–e) Comparison of conservation between RAD51 and RadA in and around the BRC4 binding site in RAD51. (c) RAD51 (surface representation) in complex with BRC4 peptide (blue tube with side chains as sticks; PDB: 1N0W) shows the BRC4 interacting residues in green on the surface. (d) Schematic map of the residues in the extended BRC4 binding site and oligomerisation groove, with RadA residues labelled in green and orange for identical or non-identical residues with RAD51, respectively, followed by RAD51 residue labels in green. Different parts of the BRC repeat and oligomerisation epitope binding sites are highlighted in grey. For orientation, the positions of the labelled binding sites are approximately in the equivalent positions in the two proteins at either side. (e) Structure of RadA ATP domain (PDB: 1PZN, chain A) bound to the oligomerisation peptide (blue tube with side chains as sticks,). The surface of RadA ATPase domain is coloured light green for identical residues with RAD51 and orange for non-identical residues. The structures of (c) RAD51 and (e) RadA are shown in the same orientation after superpositioning.