Abstract
Macrocyclization is a common feature of natural product biosynthetic pathways including the diverse family of ribosomal peptides. Microviridins are architecturally complex cyanobacterial ribosomal peptides whose members target proteases with potent reversible inhibition. The product structure is constructed by three macrocyclizations catalyzed sequentially by two members of the ATP-grasp family, a unique strategy for ribosomal peptide macrocyclization. Here, we describe the detailed structural basis for the enzyme-catalyzed macrocyclizations in the microviridin J pathway of Microcystis aeruginosa. The macrocyclases, MdnC and MdnB, interact with a conserved α-helix of the precursor peptide using a novel precursor peptide recognition mechanism. The results provide insight into the unique protein/protein interactions key to the chemistry, suggest an origin of the natural combinatorial synthesis of microviridin peptides and provide a framework for future engineering efforts to generate designed compounds.
The diverse biosynthetic pathways to natural product secondary metabolites produce enormous structural diversity and biological activity1,2. Members of the large and diverse family of ribosomally synthesized and post-translationally modified peptides (RiPPs) exhibit therapeutically useful properties including antitumor, antifungal, antibacterial and antiviral3. RiPPs are derived from short DNA-encoded precursor proteins, necessarily composed of proteinogenic amino acids. The precursor proteins generally contain an N-terminal leader peptide4,5 (or less common a C-follower peptide6), which is disposed during maturation, releasing one or more core peptides. Enzymatic post-translational modification of the core peptides installs functional/structural motifs and include, for example, dehydration, epimerization, heterocyclization, prenylation, lanthionine formation and macrocyclization4. Post-translational modification commonly requires the presence of a leader peptide, as the core peptide region alone is not recognized for modification7,8.
Genetic engineering of core peptides is an attractive and proven approach to create libraries of biomedically relevant macrocyclic peptides, and recent examples include manipulation of lasso peptides and microviridins in the heterologous host E. coli9,10. Lasso peptides, as indicated by their names, are peptides assembled as a threaded lasso and target cell surface receptors11; microviridins are depsipeptidyl toxins inhibiting serine proteases in the nM range10. Engineered, optimized heterologous expression can produce diverse RiPPs with a high yield. For example, reconstruction of the tru pathway in E. coli results in the high-yield production of diverse natural and designed cyanobactins12.
Microviridin B biosynthesis in M. aeruginosa NIES298 involves five clustered genes (mdnA-E) (Supplementary Results, Supplementary Fig. 1)13. The gene mdnA encodes a 49-amino acid precursor peptide with 14-C-terminal amino acids representing the core peptide region of the final natural product. MdnB and mdnC translate to two homologs of the ATP-grasp ligase family. This cluster also contains mdnD, encoding an N-acetyl transferase, and mdnE, encoding a putative duel-function polypeptide protease and transporter. The leader peptide of MdnA is prerequisite in the microviridin biosynthetic pathways, with a strictly conserved PFFARFL motif key to the recognition by two ATP-grasp macrocylases (Supplementary Fig. 1a)14. MdnC and MdnB condense MdnA to form a tricyclic MdnAΔ3 (MdnA-3H2O) intermediate. The order of bond formation in the three macrocyclization reactions is well resolved, typified by the investigation of microviridin K biosynthesis in Planktothrix agardhii CYA126/8 (Supplementary Fig. 1)15. The first ATP-grasp ligase (MvdD, homologous to MdnC) catalyzes the Thr/Asp and Ser/Glu lactonization, and the intermediate is then modified by MvdC (homologous to MdnB) through the Lys/Glu lactamization. Besides the PFFARFL motif, the core peptide region (MvdE) does possess limited modification tolerancy14.
On the basis of domain similarity and secondary structure prediction, the two macrocyclases in microviridin pathways are three-domain ATP-grasp proteins exemplified by RimK, a ligase involved in poly-α-glutamic acid synthesis and ribosomal protein S6 modification16. The ATP-grasp superfamily is characterized by a unique ATP-binding fold that ‘grasps’ a molecule of ATP between its C-domain and central domain17. Core peptide binding and cyclization is most likely catalyzed between the N-domain and the central domain18. Several long loop regions between the three subdomains of ATP-grasp ligases increase the overall protein flexibility and establish the basis for the recognition of structurally diverse substrates. Most ATP-grasp ligases are dimeric in structure with some examples of tetrameric, dimers of dimers. ArgX in arginine biosynthesis ligates glutamate to the carrier protein LysW19, and the tetrameric ArgX consists of two dimers interacting through the central domains and binding two LysW proteins, each sandwiched between two ArgX N-domains. Additional members of the ATP-grasp family include D-alanine:D-alanine ligase20 and diphosphoinositol pentakisphosphate kinase-221. While MdnB and MdnC produce secondary metabolite microviridins, most ATP-grasp ligases engage in the primary metabolism.
The microviridin J biosynthetic gene cluster (Fig. 1A) identified in M. aeruginosa MRC contains the genes mdnA-C, with the notable absence of mdnD and mdnE homologs (Supplementary Fig. 1)13. Mature microviridin J is a toxin that causes lethal molting disruption in Daphnia through inhibition of serine-type proteases22. Compared with other microviridins, microviridin J contains a unique Arg that facilitates its binding to trypsin10. The two ATP-grasp ligases, MdnB and MdnC, in M. aeruginosa MRC share a significant degree of sequence homology (40%) between each other, but little (< 20% sequence identity, Supplementary Fig. 2) to known crystal structures, limiting the generation of reliable homology models for either enzyme.
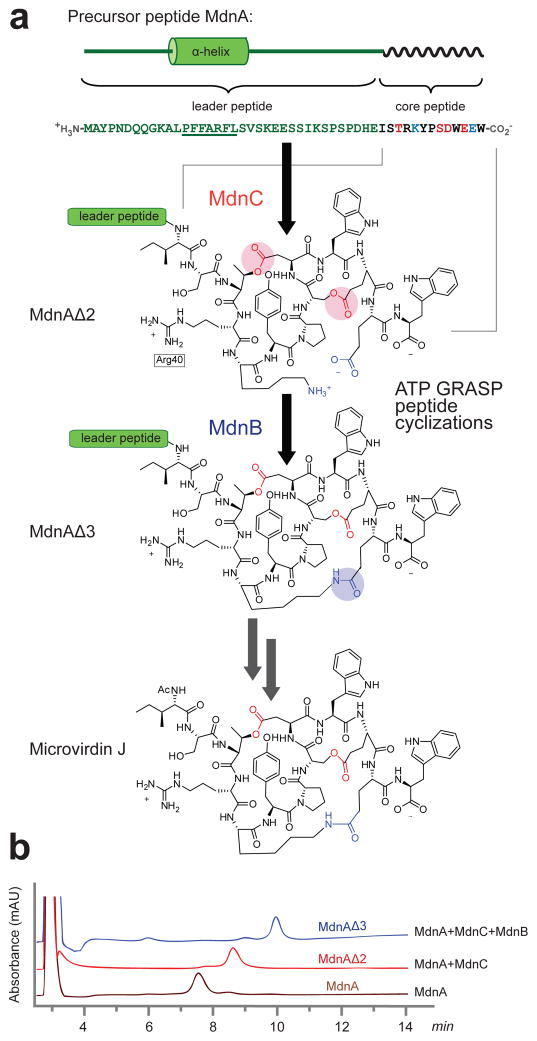
Figure 1. Biosynthesis of microviridin J.
(a) The microviridin J biosynthetic pathway in Microcystis aeruginosa MRC contains three gene products: MdnC and MdnB catalyze the macrocyclization of MdnA to generate the intermediates MdnAΔ2 and MdnAΔ3, respectively. The cleavage of leader peptide followed by an acetylation produces the mature microviridin J. The residues coupled by MdnC are colored red and likewise MdnB, blue. (b) In vitro cyclization of MdnA with purified MdnC and MdnB enzymes. HPLC analysis shows separation of the precursor peptide and two macrocyclization intermediates (wavelength=220 nm).
Cyclization of peptides is a common approach in both natural and synthetic systems to generate biologically-active peptides. The cyclic structures are less susceptible to in vivo degradation and proteolysis and present rigid conformations entropically-favored to bind targets. Several enzymatic strategies are known to catalyze the cyclization of peptides including serine protease-type mechanisms in nonribosomal23 and ribosomal peptide24,25 pathways along with ATP-dependent activation paths10,26. These approaches often exhibit high degrees of specificity and efficiency that can rival synthetic methods.
Here we describe the structure and biochemical characterization of the two peptide macrocyclases, MdnC and MdnB, from the microviridin J gene cluster. The results provide a structural basis for the interactions of peptide macrocyclases with their precursor peptide substrates, a common and integral feature of pathways to ribosomal peptide natural products.
RESULTS
MdnB and MdnC catalyze MdnA cyclization
Full-length MdnB and MdnC were heterologously expressed in E. coli as C-terminal hexa-histidine fusion proteins utilizing a short alanine linker between the tag and the enzymes. As is common with the ATP-grasp superfamily, size-exclusion chromatography profiles and native-PAGE analysis indicate that both MdnB and MdnC form homodimers (Supplementary Fig. 3). In vitro assays using purified MdnC and MdnB showed complete cyclization of MdnA to the microviridin precursor in the presence of Mg2+ and ATP. MdnC catalyzes two lactonizations to form MdnAΔ2, while MdnB installs the final macrolactam to produce MdnAΔ3 (Fig. 1, Supplementary Fig. 4 and Supplementary Table 1). Additionally, kinetic analysis of the MdnC-catalyzed double cyclization resulted in a measured KM (MdnA) of 23.8±1.2 μM and apparent kcat of 0.47±0.02 min−1 at 25°C (Supplementary Fig. 5).
MdnA leader peptide interacts with MdnB and MdnC macrocyclases
A hallmark of ribosomal peptide biosynthetic pathways is protein/protein interactions of precursor peptides with the catalytic enzymes through specific structural elements. MdnA is 49 amino acids in length with 13 of those amino acids included in the final natural product structure. To understand the interaction of MdnA with the macrocyclases, we conducted ITC-based binding assays. Full-length MdnA (amino acids 1–49, Fig. 2, i) binds MdnC with a measured binding constant (KD) of 112±52 nM (Supplementary Fig. 6 and Supplementary Table 2). This binding interaction is ATP-independent and the addition of the stable ATP analog AMPPNP, or ADP, does not change the MdnA/MdnC interaction thermal profile. We next removed the core peptide region of MdnA to create a leader peptide-only analog (MdnA1–35, ii), and observed the similar level of interactions between MdnA1–35 and MdnC (KD = 146±52 nM). These results indicate that the MdnA/MdnC interaction is predominantly leader peptide-dependent, while the core peptide of MdnA (MdnA36–49, vi) does not contribute significantly in the initial stage of substrate recognition. Using a similar approach, we observed a weaker interaction between MdnB and full length MdnA or MdnA1–35 (KD values of 4.8±1.0 and 2.8±0.8 μM, respectively), not an unexpected result as the natural substrate for MdnB is the dicyclic-MdnAΔ2 and not MdnA. In order to further test the binding of leader peptide to the second cyclase, MdnB, the intermediate MdnAΔ2 was prepared and the binding affinity measured, resulting in a KD value of 6.4±2.0 μM. This result indicates that the MdnAΔ2/MdnB interaction is leader peptide-driven following the common logic of RiPPs.
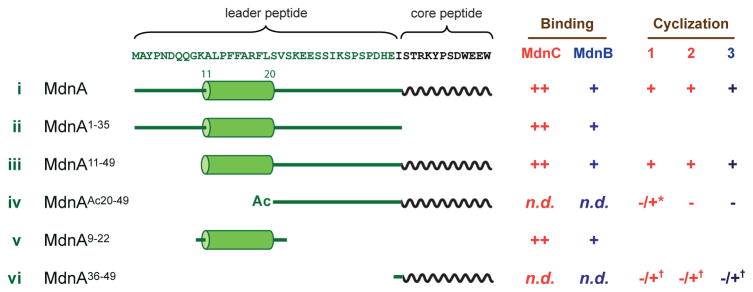
Figure 2. Binding and cyclization activity of macrocyclases toward MdnA variants.
Truncated MdnA variants were utilized to probe the binding and activity determinants of MdnC and MdnB. For binding, ++ represents strong binding and +, weak binding (see Supplementary Fig. 6 and Table 2). Cyclization is indicated by a ‘+’ or ‘−’ for no observed reaction. n.d. = no activity detected. ‘*’ indicates in trans reaction with MdnA1–35 and ‘†’ in trans reaction with MdnA9–22.
Additionally, we determined the role of the strictly conserved region of the MdnA leader peptide, PFFARFL (Fig. 2). An N-terminal truncated MdnA (MdnA11–49, iii) containing PFFARFL along with flanking amino acids binds to both enzymes and is cyclized. In contrast, removal of the conserved region (MdnAAc20–49, iv) results in a peptide that neither binds nor is processed. A minimal peptide containing the conserved region (MdnA9–22, v) was then tested for binding with both MdnC and MdnB, resulting in KD values of 308±62 nM and 1.55±0.25 μM respectively. The values are comparable to the MdnA/MdnC, and MdnA/MdnB interactions, suggesting that the strictly conserved region is the main driving force in substrate recognition.
Overall structures of MdnC and MdnB
To further understand the leader peptide prerequisite in the microviridin biosynthetic pathway, we determined the crystal structures of MdnC and MdnB at resolutions of 2.7 Å and 2.3 Å, respectively. The phase information for both MdnC and MdnB was calculated from SeMet-derivatized proteins using SAD (Supplementary Table 3). Multiple crystallization trials of MdnC and MdnB with MdnA and/or ATP/nonhydrolyzable analogs all resulted in the same “active site free” structures, with several unresolved isomorphous loop regions key to the enzymatic reactions and no substrate or cofactor apparent in the electron density maps. To obtain a more complete model, MdnA1–35 was chosen as the alternative substrate mimic to probe the precursor peptide/enzyme interactions. MdnC was crystallized in the presence of the C-truncated MdnA1–35 (without the core peptide region) and the structure of the complex was solved. The MdnC/MdnA1–35 complex was in a tetragonal form, with four dimers per asymmetric unit. In contrast, only the apo-form of MdnB was obtained when co-crystallized with MdnA1–35. The orthorhombic crystal form of MdnB consists of two protomers per asymmetric unit, where the biological unit (dimer), is formed via crystallographic symmetry. Compared with the circular octameric geometry of MdnC in the crystal, MdnB dimers pack in a linear form (Fig. 3 and Supplementary Fig. 7). Residues 2–244/250–325 in MdnC (chain A) and 5–238/254–326 in MdnB (chain A) were successfully built into the electron density maps. Residues 11–22 of MdnA (chain I) are resolved in the MdnC/MdnA1–35 complex.
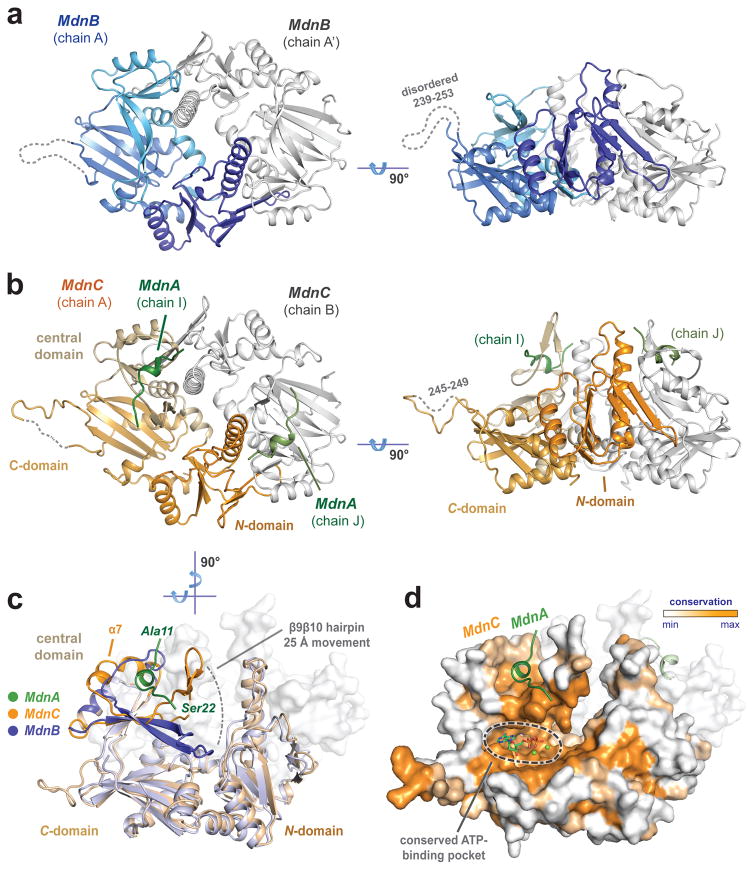
Figure 3. Overall structures of MdnC and MdnB.
(a) Two views of MdnB (blue/grey) and (b) MdnC (orange/grey) dimers. MdnC forms a co-complex with the precursor peptide MdnA1–35 (green). (c) Superimposition of the MdnC and MdnB protomers illustrate domain movement involved in precursor peptide binding. (d) Evolutionary conservation map of MdnC protomer highlights conserved regions of the C-domain. The modelled ADP/Mg2+ binding site is noted on the map.
Both MdnB and MdnC fit in the overall ATP-grasp ligase structural motif, with three subdomains: N-domain, central domain and C-domain (Fig. 3). MdnC and MdnB form similar homodimer assemblies with an interface area of approximately 2748 Å2 (MdnC) and 2442 Å2 (MdnB), respectively. Comparison of the dimers of MdnC and MdnB show an overall Cα RMSD of 1.7 Å. Amino acids near the dimeric interface are predominantly hydrophobic in nature, while the ATP binding sites have a higher calculated electrostatic potential (Supplementary Fig. 8). The dimeric interaction is mediated through two key antiparallel β-strands between the N-domain (β3) and the neighboring central domain (β′8) (Supplementary Fig. 9). These two strands are structurally conserved in all ATP-grasp ligase family members. In addition, α3 extends along the neighboring central domain, interacting with β′9 and contributing to the stabilization of the dimer. Compared with other reported ATP-grasp ligases, the 23-amino acid long helix (α3) in MdnC/B is atypical, as others commonly contain a much shorter helix at this position. Two helices, α4 and α′4, connecting the N-domain and central domain, are located in the middle of the dimeric interface. The α4 and α′4 in MdnC dimers are in a close contact, whereas in the MdnB dimer they are ~6 Å apart, creating a top-to-bottom tunnel-like feature (Supplementary Fig. 8).
The MdnC and MdnB protomers are overall structurally similar, but there are several differences in their central domains (Fig. 3c), specifically in the β9β10 hairpin region and the α7 region (Supplementary Fig. 10). MdnC possesses a long two-stranded antiparallel β-sheet (β9β10), followed by a relatively ordered helix, α7. The β9β10 and α7 regions are anchored by the MdnA leader peptide. On the contrary, the β9β10 hairpin of MdnB lays on the C-domain, while the α7 region is disordered into a flexible loop. In the two structures, the β9β10 hairpins are in significantly different conformations; the turns of the hairpins are 25 Å apart comparing MdnC and MdnB.
Key residues for nucleotide binding and catalysis
The macrolactone cyclizations of Thr39/Asp45, Ser43/Glu47 (by MdnC) and Lys41/Glu48 (by MdnB) require Mg2+ and ATP15. The reactions of MdnC and MdnB both generate ADP, but not AMP (Supplementary Fig. 11a). These results suggest that the cyclizations are most likely to occur via acylphosphorylation, as opposed to (also common) the generation of acyl-AMP intermediates. Unfortunately, as mentioned and despite extensive efforts, we were unable to observe ATP, ADP or various non-hydrolyzable analogs soaked or co-crystallized with MdnC or MdnB in the calculated electron density maps. Also, unlike other ATP-grasp ligases, whose interactions with ATP have been experimentally determined19, neither MdnC nor MdnB show an observable heat change with ATP (and its analogs) using ITC analysis. Nonetheless, the ATP binding pocket of the ATP-grasp ligases is structurally conserved, allowing the determination of the key residues in MdnC and MdnB that interact with the nucleotide and facilitate the macrocyclizations (Fig. 4a and Supplementary Table 4). Using structural alignment, critical residues involved in ATP interactions and metal ion coordination include Lys125, Lys166, Gln207, Glu215, Asp281, Glu294 and Asn296 (for MdnC). To probe the importance of these residues in the MdnC reaction, we mutated each to alanine. The mutation of any of these residues (K125A, K166A, Q207A, D281A and E294A/N296A, Supplementary Fig. 11b) completely abolished the MdnC cyclization activity. These residues are predicted to be primarily involved in ATP binding and not interaction with the precursor peptide. Indeed, representatives of these MdnC mutants (D281A and E294A/N296A) showed the similar level of interaction with the precursor peptide MdnA as measured with ITC (Supplementary Fig. 6).
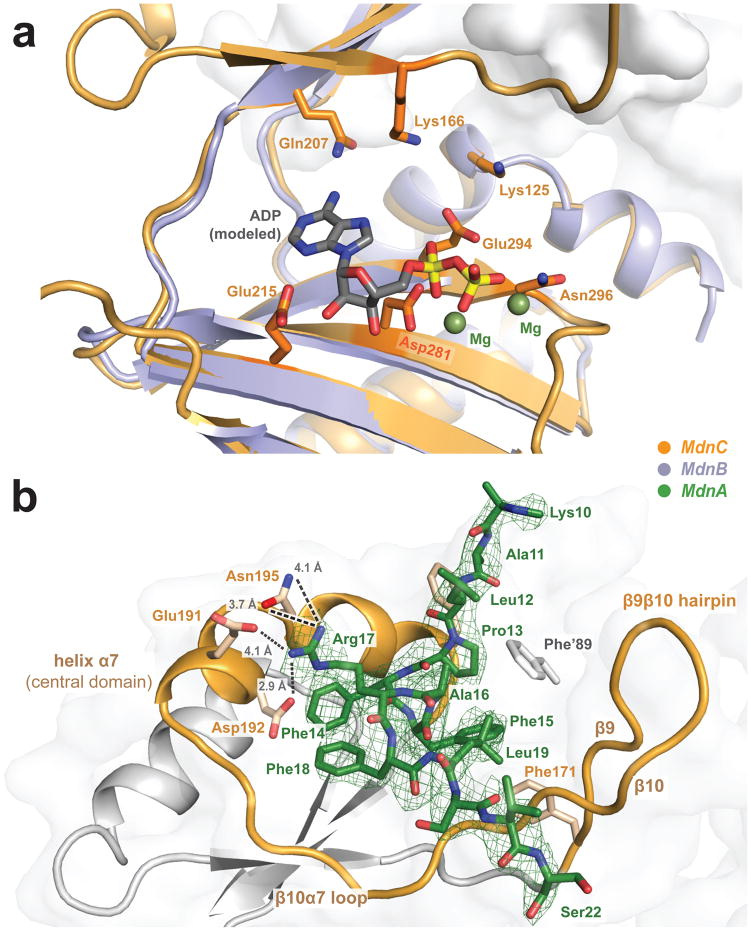
Figure 4. MdnC uses ATP to catalyze the macrocyclization of the precursor peptide MdnA.
(a) Modelled ADP interacts with the C-domain and central domain of MdnC. The positions of Mg2+ and ADP are adapted from the aligned LysX structure (PDB entry 3VPD, 20% sequence identity). (b) Interactions of MdnC with the precursor peptide MdnA. The strictly conserved region of MdnA (PFFARFL) is well-resolved in the electron density map. Helix α7 and β9β10 hairpin of MdnC sandwich the helical region of the resolved MdnA leader peptide. Calculated mFo-DFc map is displayed at a 1.5σ contour level.
Recognition of MdnA leader peptide by MdnC and MdnB
Our binding measurements suggest that MdnA interacts with MdnC in a core peptide-independent manner (Supplementary Fig. 6 and Supplementary Table 2). Consistent with this, amino acids 11–22 of MdnA1–35 were well-resolved in the calculated electron density map of the MdnC/MdnA1–35 complex. This region of the leader peptide includes the strictly conserved motif of PFFARFL bound in an α-helix conformation (Fig. 4b)14. MdnA1–35 interacts with the central domain of MdnC through electrostatic interactions between Arg17 of MdnA and Glu191/Asp192/Asn195 of MdnC α7. Compared with results from single and double mutants of the PFFARFL motif, the Arg is a key residue in the MdnA conserved region. In the related system, the homologous R17N mutant (M. aeruginosa NIES843, microviridin L) showed decreased dicyclic- microviridin production14. In most other MdnC-like macrocyclases, Glu191 is replaced by an aspartate residue, while Asp192 is well conserved among all microviridin macrocyclases (Supplementary Fig. 12). Meanwhile, electrostatic interactions between the backbone amides of Ser20 (MdnA) and Val182 (MdnC) induce the movement of MdnC β9β10 hairpin toward MdnA1–35. The interface area of bound MdnA1–35 and MdnC is ~740 Å2.
To probe the key residues of MdnC for MdnA binding, we created two MdnC variants with double mutations, MdnCE191K/D192K and MdnCE191A/D192A (referred to as MdnCKK and MdnCAA, respectively). The charge reversal of the conserved Glu191 and Asp192 residues completely abolished the MdnA/MdnCKK interaction (Supplementary Fig. 6), and MdnCKK had no macrocyclization activity toward the full length MdnA. On the other hand, the more conservative mutant MdnCAA retained partial cyclization activity but had no detectable MdnA/MdnCAA binding interaction via ITC analysis. Compared with WT MdnC, MdnCAA exhibited a significantly slower reaction, and promoted the formation of MdnAΔ1 and MdnAΔ2 in a ~1:1 ratio (Supplementary Fig. 11). These observations confirmed the roles of Glu191 and Asp192 and α7 in precursor protein recognition.
To further evaluate the importance of observed MdnA structural elements in the enzymatic macrocyclization chemistry, we created three truncated precursor protein variants and characterized their ability to be cyclized (Fig. 2). MdnA36–49 (vi) does not include the leader peptide region, MdnAAc20–49 (iv) is missing just the conserved, structurally ordered α-helix region, while MdnA11–49 (iii) contains the α-helical PFFARFL motif but not the N-terminal ten amino acids of MdnA, which were not observed in the MdnC/MdnA1–35 co-complex structure. As predicted, MdnAAc20–49 and MdnA36–49 showed no binding to MdnC and were not cyclized in our in vitro assays (Supplementary Fig. 13bc). In contrast, the MdnA11–49 variant bound to MdnC with comparable parameters as full length MdnA and could be dicyclized (Supplementary Fig. 6 and Supplementary Fig. 13a). Additionally, the reaction product MdnA11–49Δ2 was further converted into MdnA11–49Δ3 by MdnB. These results along with the crystal structure have revealed the minimum requirements for efficient precursor peptide binding and cyclization.
Next, we explored the potential of leader-free macrocyclization following similar approaches used in other RiPP biosynthetic pathways27,28. We first attempted in trans cyclization of an ‘inactive’ substrate (MdnAAc20–49) with the MdnA leader peptide MdnA1–35. Indeed, the combination of ‘inactive’ MdnAAc20–49 with the ‘inducer’ MdnA1–35 led to the production of a cyclic intermediate MdnAAc20–49Δ1, but unexpectedly, the product of the second cyclization was not detected. Identical results were obtained employing extended reaction times or increased MdnA1–35 concentration. MdnAAc20–49Δ1 was not a substrate of MdnB in the presence or absence of inducer MdnA1–35 (Supplementary Fig. 13b). In order to further delineate the minimal leader and core peptides regions, we applied MdnA9–22/MdnA36–49 (Fig. 2, v and vi) in an in trans reaction with MdnC/MdnB. The results show that this small peptide, MdnA36–49, can be fully cyclized to generate MdnA36–49Δ3 only in the presence of conserved α-helical peptide MdnA9–22 (Supplementary Fig. 13c).
DISCUSSION
The microviridin J gene cluster of M. aeruginosa MRC contains a minimal set of genes (mdnA-C) for the biosynthesis of microviridin, making it an excellent model system for the study of RiPP biosynthesis and cyclization. We demonstrated the in vitro catalytic activity of MdnC and MdnB in the microviridin J biosynthetic pathway, and structurally characterized the two macrocyclases along with their precursor protein MdnA. Both MdnC and MdnB macrocyclases have several long interdomain loops that show significant disorders in the electron density maps. Similar to many ATP-grasp family members, this feature likely provides the interdomain flexibility necessary for the binding of the 13-amino-acid core peptide substrate.
MdnC catalyzes the first step in the post-translational modification of microviridin J by the formation of two macrolactone rings (Fig. 1, MdnA to MdnAΔ2). Compared with the ‘apo-form’ of MdnB, MdnA-bound MdnC exhibits a large movement of the central domain. The ~25 Å shift of the β9β10 hairpin region in MdnC opens the interdomain pocket, and allows the core peptide to access bound ATP between the central domain and C-domain. Meanwhile, the crystal structure of MdnB represents the ‘closed’ conformation with the β9β10 region blocking the pocket. Binding of the leader peptide of MdnA restrains the β9β10 hairpin and the α7 helix in MdnC. These regions may act as allosteric sites for subsequent core peptide recognition and catalysis, assisting in orienting the inherently disordered active site. In support of this hypothesis, mutagenesis of conserved residues on α7 significantly alters the enzymatic efficiency.
The structural basis and mechanism of precursor peptide recognition is the key factor in enzyme/substrate selectivity in RiPP pathways. Only limited structures of RiPP biosynthetic enzymes have been determined in complex with their corresponding precursor peptides, and include dehydratase NisB involved in nisin biosynthesis29 and heterocyclase, LynD, involved in cyanobactin biosynthesis28. Both enzymes use a three-stranded antiparallel β-sheet to interact with the conserved motif of the leader region of the precursor peptide. A RiPP precursor peptide recognition element (RRE) has been predicted based on these partial co-complex structures and related mutagenesis studies5,30,31. By comparison, MdnC and MdnB do not possess a similar RRE. The interaction between MdnA and the two ATP-grasp ligases represents a novel type of RiPP precursor peptide recognition mechanism. Additional reported leader peptides tend to contain α-helices in solution, and presumably maintain this structure when bound to the biosynthetic proteins4. Structural comparison between MdnA-bound MdnC and apo-MdnB indicates that the α-helical leader peptide is critical for the conformation change of the biosynthetic enzymes, while the conformational change is the key factor modulating the enzyme activity. While the α-helix is important for post-translational modification machinery, the first 10 amino acids at the N-terminus of MdnA (1–10) are not required for precursor peptide recognition. Similar substrate specificity has been observed in the lacticin biosynthetic pathway, where a precursor peptide LctA variant with the deletion of its N-terminal eight amino acids could be fully processed32. Another example of N-terminal dispensability of the N-terminus of leader peptides is the biosynthesis of lasso peptide microcin J25, in which the first 25 of the 37 residues in the leader peptide could be removed33. Additionally, compared with MdnB, the MdnC/leader peptide (MdnA1–35, and MdnA9–22) binding interaction is ~10 fold tighter. The differential binding affinities of the precursor peptides can be rationalized in several ways. It is reasonable to predict that the linear MdnA peptide is less stable and thus is required for it to be processed rapidly to the macrocycle.
In trans activation of the macrocyclase by the helical leader peptide alone led to an active enzyme, an interesting feature of several RiPP systems33,34. In trans processing of the truncated MdnAAc20–49 (iv, without the conserved PFFARFL leader motif) with added MdnA1–35 by MdnC produced a ‘trapped’ product with a single cyclization. However, utilizing the minimal leader peptide (MdnA9–22, v) along with a minimal core peptide (MdnA36–49, vi) allowed vi to be fully cyclized to MdnA36–49Δ3 with MdnC and MdnB whereas no cyclization occurred without v. The reaction is comparable to LctM, shown to catalyze multiple dehydrations of leader-free LctA with a high concentration of in trans leader peptide27.
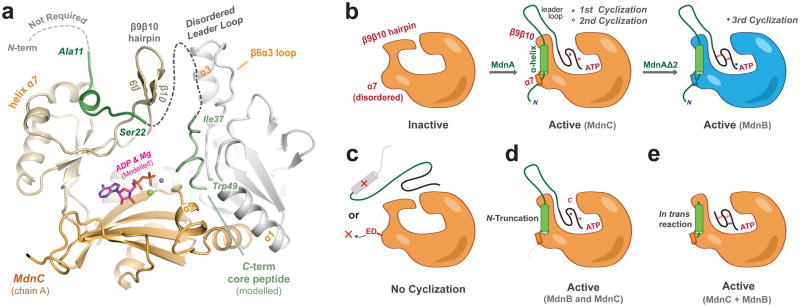
The helix of the MdnA leader peptide is ~15 amino acids away from the core peptide of MdnA. This peptide region was unresolved in the MdnC/MdnA1–35 complex structure, and may be disordered and/or flexible. However, simple modeling suggests the antiparallel β-strands (β9β10) between the leader peptide and the ATP active site could interact in a β-sheet motif, similarly to the PatE/LynD interaction28. The MdnA α-helical C-terminus might extend along MdnC β10 near the MdnC dimeric interface, and then fold back to the active site between MdnC N-domain/central domain, where the core peptide could approach bound ATP for activation (Fig. 5a). Another member of the ATP-grasp ligase, ArgX uses the N-domain to interact with the β-barrel of LysW, and the C-terminus of LysW extends into the ArgX active site19. In contrast, the N-domain of both MdnC and MdnB show significant differences compared with the ArgX N-domain (Supplementary Fig. 2). One of the structural differences between MdnB and MdnC is the tunnel-like feature of MdnB at its dimeric interface. This larger central cavity could be necessary for binding and processing of its more rigid substrate, MdnAΔ2.
Figure 5. Leader peptide directed peptide macrocyclization in the microviridin J biosynthetic pathway.
(a) Model of the interactions of full length MdnA (green) with the macrocyclase, MdnC. The cyclized core peptide region was adapted from pdb entry 4KTU and docked into MdnC with modelled ADP. (b) The leader peptide of MdnA activates macrocyclation by orienting α7 and by inducing a large shift of the β9β10 hairpin. (c) The α-helical element of the precursor peptide, as well as residues on α7 (Glu191/Asp192, ‘ED’), are critical and removing these elements result in no cyclization, (d) whereas the N-terminus of the substrate is not requisite for either MdnB or MdnC macrocyclizations. (e) In trans activation of MdnC using the α-helical element of the precursor peptide and an inactive substrate produces fully active macrocyclases.
We were not able to observe a complex structure of MdnC or MdnB with the MdnA core peptide (the region cyclized and present in the final product), nor did we observe any core peptide binding interaction with MdnC or MdnB by ITC. This suggests that the macrocyclase-core peptide interaction is not the key determinant for binding/recognition and the interaction between the macrocyclase and the leader peptide plays a more significant role. Indeed, several reports have shown that alteration of the core peptide region is tolerated whereas changes to the conserved region of the leader peptide are not14,34. This is also consistent with the natural, genetic variability observed in microviridin gene clusters. In the P. agardhii gene cluster, mvdE and mvdF represent two distinct precursor proteins that are both substrates for the macrocyclases (Supplementary Fig. 1). Additionally, there are several examples of a single precursor peptide gene product containing one leader peptide and multiple core peptides. The all7013 gene product in Nostoc sp. PCC 7120 contains the strictly conserved α-helical element in the leader peptide region and three different core peptide regions, indicating the potential to form three different final products (Supplementary Fig. 1). Similar combinatorial assembly in RiPP systems has been detailed in the patellamide biosynthetic pathway35.
The structural basis of substrate precursor peptide binding in the microviridin biosynthetic pathway is summarized in Fig. 5. The α-helical region of the leader peptide binds to the α7 of macrocyclases and induces a conformational change of the β9β10 hairpin, which allows the core peptide region to access the ATP binding pocket for activation and cyclization. These processes require the presence of the helix element on the leader peptide, but the amino acids N-terminal to this helix region are not required for the macrocyclizations. The disruption of leader peptide/macrocyclase interaction by mutating the key residues on α7 also abolishes the cyclization reactions. In addition, fully activated macrocyclizations can be observed through in trans reactions. The results presented provide novel insights into the understanding of RiPP biosynthetic pathways and facilitate their discovery and engineering
ONLINE METHODS
Protein cloning, expression and purification
Codon-optimized, full-length mdnB and mdnC (M. aeruginosa MRC) were purchased from Mr. Gene (Regenburg, Germany) supplied in pMA vectors. mdnB and mdnC were amplified using PCR (Supplementary Table 5) and a short C-terminal AAAHHHHHH hexa-histidine tag was encoded in the C-terminal primers bearing the Xhol restriction site for both MdnB and MdnC. The mdnB and mdnC genes were then ligated into the expression vector pET30a. MdnC was expressed in E. coli BL21(DE3) pLysS. Cultures (1 L) were grown at 37°C to an OD600=0.6 and overexpression was initiated by adding isopropyl β-D-1-thiogalactopyranoside (IPTG, final concentration 100 μM). Growth was continued for 12 h at 18°C, before the cells were harvested by centrifugation. Cell pellets were resuspended in 25 mL of 0.5 M NaCl and 20 mM Tris-HCl, pH 7.5, and lysed at 14,000 psi through a nitrogen-pressure microfluidizer cell (M-110L Pneumatic). The lysate was clarified by centrifugation at 15,000g for 20 min at 4°C. MdnC was purified by immobilized metal affinity chromatography (HisPur Ni-NTA Resin, Thermo Scientific). After binding for 1 h, the resin was washed with 4X 10 mL of 0.5 M NaCl, 10 mM imidazole and 20 mM Tris-HCl, pH 7.5, and the bound protein was eluted with 3X 2 mL of 0.5 M NaCl, 250 mM imidazole and 20 mM Tris-HCl, pH 7.5. The elution fraction was dialysed into a low salt buffer (50 mM NaCl and 20 mM Tris-HCl, pH 7.5), and further purified by anion exchange chromatography (MonoQ HR 10/10, AKTA FPLC System, GE Healthcare) with a linear gradient of 50–500 mM NaCl over 30 min, followed by a size-exclusion chromatography (HiLoad 16/60 SuperDex-200 column, AKTA FPLC System, GE Healthcare) with buffer 150 mM NaCl, and 20 mM Tris-HCl, pH 7.5. Pooled MdnC was concentrated to ~10 mg mL−1 for crystallization. MdnB was overexpressed and purified following the same procedure described for MdnC.
MdnC mutagenesis
Mutations of MdnC were carried out using the Q5 Site-Directed Mutagenesis Kit (NEB) following the manufacturer’s instructions. The validity of mutagenesis was confirmed by DNA sequencing. The MdnC mutant list and primers designed for the mutagenesis are listed in Supplementary Table 5.
Designed MdnA variants
Full-length MdnA (residues1–49) (Genscript, USA and SynPeptide), MdnA1–35 (SynPeptide), MdnA11–49 (LifeTein) and MdnAAc20–49 (EZBioLabs) were purchased from custom peptide suppliers. MdnA9–22 and MdnA36–49 were synthesized following standard protocols (Synergy 432A peptide synthesizer, FMOC chemistry/Wang resin, reagents Sigma-Aldrich). MdnA9–22 contains the highly conserved region of the leader peptide, while MdnA36–49 is the core peptide region of MdnA.
Macrocyclization of MdnA variants
For MdnA (and its variants) cyclization in vitro, MdnA substrates were treated with 100 mM Tris-HCl, pH 8.0, 2 mM ATP, 10 mM MgCl2, 50 mM KCl, and 1.8 μM MdnC (or 1.8 μM MdnC added with 1.8 μM MdnB) at 37°C and quenched after 4 h with equal volumes of 0.5 M EDTA. HPLC was used for product analysis: 30% acetonitrile for 3 min, and 30–40 % acetonitrile from 3–14 min in 0.1 % TFA-water. LC-MS was used to analyze the corresponding cyclization with the method: 3% 0.5% FA-acetonitrile for 3 min, and 3–98% 0.5% FA-acetonitrile from 5–11 min in 0.5 % FA-water.
MdnC-MdnA dicyclization kinetics
The kinetics of MdnC and MdnA dicyclization reaction were analyzed using an HPLC based assay. Various concentrations of MdnA (8.75 – 52.5 μM) were processed with 1.8 μM MdnC with the reaction conditions described above at 25°C, and quenched at various time points (2–60 min). The reaction mixtures were analyzed with analytical HPLC equipped with an auto-sampler and the method: 30 % acetonitrile for 5 min, and 30–40 % acetonitrile from 5–11 min in 0.1 % TFA-water. The linear form of MdnA eluted at 8.5 min, and the MdnC reaction product, dicyclic-MdnAΔ2 (MdnAΔ2) eluted at 9.5 min. LC-MS analysis verified that the product was 35.9 Da difference from the standard as described for microviridin K34, indicate two cyclizations had occurred. Kinetic parameters were deduced from the integration of the starting material and product peaks areas. Reactions at each MdnA initial concentration were measured in triplicate. Error bars indicated in all figures are standard deviations (± s.d.).
Leader peptide binding affinity determination
Isothermal titration calorimetry (ITC) was used to determine the MdnB, MdnC and MdnA interactions. 200 μL of 0.1 mM enzyme in 50 mM KCl, 10 mM MgCl2 and 100 mM HEPES-NaOH, pH 8.0 was placed in a MicroCal iTC200 reaction cell, while 39.6 μL full (or partial) length MdnA peptide (1.0 mM) was titrated into the protein solution over time at 25°C. Heat change during the reaction was detected and recorded for binding affinity calculation. All titrations were repeated in triplicate. The binding parameters and corresponding error are calculated using the Origin software package36.
Preparation of SeMet labelled protein
Selenomethionine (SeMet) labelled MdnC was produced using methionine auxotroph E. coli (T7 Crystal Express, NEB). To improve the crystal diffraction quality of SeMet-MdnC, mutagenesis was carried out to decrease the number of nonconserved Met (M25K, M104L, M167L, M181N, and M282I). Cells were cultured in 6 L M9 minimal media supplemented with 50 mg L−1 L-Met and grown to an OD600=0.6 at 37°C. The cells were then pelleted by centrifugation, resuspended in 6 L of fresh M9, and incubated at 37°C for 3 h to deplete the intercellular methionine. 50 mg L−1 L-SeMet was then added to the culture 20 min before the overexpression, which was initiated by adding 100 μM IPTG followed by growth for 16 h at 18°C. The purification for SeMet labelled MdnC was the same as described above for the native protein with the addition of 10 mM dithiothreitol (DTT) to the dialysis, anion exchange, and gel filtration purification buffers. SeMet labelled MdnB was prepared and purified in the same fashion.
Crystallization, data collection and crystallographic analysis
Purified MdnC was mixed with 10 eq. of AMP-PNP, 1.2 eq. of MgCl2 and 1.05 eq. of MdnA1–35 (final concentration [MdnC] = 7.4 mg mL−1) and allowed to equilibrate at 4°C overnight. Initial crystal screening was performed in a vapor diffusion, sitting drop format using commercial sparse matrix screens. Small plate clusters were identified in a condition containing 200 mM ammonium sulfate, 20% PEG-3350, and 100 mM bis-Tris-HCl, pH 6.5. Optimization of salt and pH along with microseeding were performed in hanging drop format at 20°C.The resultant crystals with a maximum size of ~50×200×200 μm were obtained in a final condition that contained 250 mM ammonium sulfate, 28% PEG-3,350, 8% dioxane and 100 mM bis-Tris-HCl, pH 5.5.
MdnB was concentrated to 6.0 mg mL−1 and premixed with 5 eq. AMP-PNP and 1.5 eq. MgCl2. The mixture was incubated at 4°C for 4 h and screened with commercial sparse matrix in a vapor diffusion, sitting drop format. Diffracted plate-shaped single crystals with a size of ~40×100×100 μm were obtained in the best condition contains 20% PEG 4,000, 20% 2-propanol and 100 mM sodium citrate, pH 5.6. SeMet labelled MdnB was crystallized in a same manner, crystals were harvested and flash frozen, with 15% v/v glycerol as cryoprotectant.
Diffraction data for MdnB crystal was collected on beamline 22-ID of the Advanced Photon Source-Argonne National Laboratory (APS-ANL) at a wavelength of 0.9787 Å. MdnC diffraction data were collected on beamline 21-ID-G of the Life Sciences Collaborative Access Team (LS-CAT) facility, APS-ANL, at a wavelength of 0.9786 Å. Data were collected at 100 K, integrated, merged and scaled using XDS package37. The MdnB SeMet-SAD data set was processed into the space group C2 with two molecules per asymmetric unit. Phases were calculated with PHENIX.AUTOSOL38,39 and the resultant partial model was used in molecular replacement for native MdnB data. PHENIX.AUTOBUILD succeeded in placing 80% of the amino acids in MdnB sequence. The remaining residues were built into the electron density maps manually with COOT40. MdnC crystals diffracted X-ray in space group P41, with 8 monomers per asymmetric unit. MdnC structure was solved with dimeric MdnB as molecular replacement model with PHASER41. PHENIX.AUTOBUILD placed 85% of the amino acids in MdnC sequence, while the rest of MdnC as well as MdnA fragments were built manually with COOT. SeMet labelled MdnC data set facilitated the phase improvement throughout the model building. Structures were refined using REFMAC542 and PHENIX.REFINE. Sigma-A weighted, simulated annealing composite omit maps were used to judge and verify structures throughout refinement. The refined MdnC and MdnB structures include Ramachandran favored (outliers) as 95% (0.6%) and 96% (0.9%), respectively. We also processed apo-MdnC crystals (without added MdnA or variants). However, the apo structure has multiple disordered regions and only ~75% of the protein sequence can be assigned into the density maps. Specifically, the MdnA leader peptide binding region (central domain) was not resolved and the structure could be refined to an Rfree value ~35%.
Crystallographic data and refinement statistics are shown in Supplementary Table 3. Sequence alignments were carried with Clustal Omega43. MdnC sequence evolutionary conservation was calculated from ConSurf44. Protein surface electrostatics were calculated and mapped with APBS45 and PDB2PQR46. Docking and analysis were performed with AutoDock 447. Structural illustrations were prepared with PyMOL (https://www.pymol.org).
Supplementary Material
Acknowledgments
We thank the staff on the 21-ID-G and 22-ID beamlines at Argonne National Laboratory, Advanced Light Source for help with data acquisition and processing. We thank T. Montavon and S. Lagishetty for providing experimental contributions supporting this work. We also are grateful to Prof. N. Polfer and A. Patrick for assistance with peptide synthesis. This work was supported by NIH GM086570 and the funds from the University of Florida (S.D.B.).
Footnotes
Accession codes. RCSB Protein Data Bank: Crystallographic data are listed under accession codes 5IG8 (MdnB), and 5IG9 (MdnC bound with MdnA).
Author contributions
K.L., H.L.C., and S.D.B. conceived the project and designed the experiments. K.L. and H.L.C. performed cloning, expression screening, protein purification, crystallization and X-ray diffraction data collection, structure solution and refinement. H.L.C. carried kinetic analysis and K.L. and G.L. prepared and analyzed mutant proteins. K.L., H.L.C., Y.D. and S.D.B. interpreted the data and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 2.Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov. 2015;14:111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 3.McIntosh JA, Donia MS, Schmidt EW. Ribosomal peptide natural products: bridging the ribosomal and nonribosomal worlds. Nat Prod Rep. 2009;26:537–559. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortega MA, van der Donk WA. New insights into the biosynthetic logic of ribosomally synthesized and post-translationally modified peptide natural products. Cell Chem Biol. 2016;23:31–44. doi: 10.1016/j.chembiol.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crone WJK, Leeper FJ, Truman AW. Identification and characterisation of the gene cluster for the anti-MRSA antibiotic bottromycin: expanding the biosynthetic diversity of ribosomal peptides. Chem Sci. 2012;3:3516. [Google Scholar]
- 7.Xie L, et al. Lacticin 481: in vitro reconstitution of lantibiotic synthetase activity. Science. 2004;303:679–681. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
- 8.Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat Chem Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan SJ, Link AJ. Sequence diversity in the lasso peptide framework: discovery of functional microcin J25 variants with multiple amino acid substitutions. J Am Chem Soc. 2011;133:5016–5023. doi: 10.1021/ja1109634. [DOI] [PubMed] [Google Scholar]
- 10.Weiz AR, et al. Harnessing the evolvability of tricyclic microviridins to dissect protease-inhibitor interactions. Angew Chemie - Int Ed. 2014;53:3735–3738. doi: 10.1002/anie.201309721. [DOI] [PubMed] [Google Scholar]
- 11.Mathavan I, et al. Structural basis for hijacking siderophore receptors by antimicrobial lasso peptides. Nat Chem Biol. 2014;10:340–342. doi: 10.1038/nchembio.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tianero MD, et al. Metabolic model for diversity-generating biosynthesis. Proc Natl Acad Sci USA. 2016;113:1772–1777. doi: 10.1073/pnas.1525438113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziemert N, Ishida K, Liaimer A, Hertweck C, Dittmann E. Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angew Chemie - Int Ed. 2008;47:7756–7759. doi: 10.1002/anie.200802730. [DOI] [PubMed] [Google Scholar]
- 14.Weiz AR, et al. Leader peptide and a membrane protein scaffold guide the biosynthesis of the tricyclic peptide microviridin. Chem Biol. 2011;18:1413–1421. doi: 10.1016/j.chembiol.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Philmus B, Christiansen G, Yoshida WY, Hemscheidt TK. Post-translational modification in microviridin biosynthesis. ChemBioChem. 2008;9:3066–3073. doi: 10.1002/cbic.200800560. [DOI] [PubMed] [Google Scholar]
- 16.Zhao G, et al. Structure and function of Escherichia coli RimK, an ATP-grasp fold, L-glutamyl ligase enzyme. Proteins Struct Funct Bioinforma. 2013;81:1847–1854. doi: 10.1002/prot.24311. [DOI] [PubMed] [Google Scholar]
- 17.Iyer LM, Abhiman S, Maxwell Burroughs A, Aravind L. Amidoligases with ATP-grasp, glutamine synthetase-like and acetyltransferase-like domains: synthesis of novel metabolites and peptide modifications of proteins. Mol Biosyst. 2009;5:1636–60. doi: 10.1039/b917682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fawaz MV, Topper ME, Firestine SM. The ATP-grasp enzymes. Bioorg Chem. 2011;39:185–191. doi: 10.1016/j.bioorg.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouchi T, et al. Lysine and arginine biosyntheses mediated by a common carrier protein in Sulfolobus. Nat Chem Biol. 2013;9:277–283. doi: 10.1038/nchembio.1200. [DOI] [PubMed] [Google Scholar]
- 20.Liu S, et al. Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies. Proc Natl Acad Sci USA. 2006;103:15178–15183. doi: 10.1073/pnas.0604905103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Falck JR, Hall TMT, Shears SB. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat Chem Biol. 2012;8:111–6. doi: 10.1038/nchembio.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohrlack T, Christoffersen K, Kaebernick M, Neilan BA. Cyanobacterial protease inhibitor microviridin J causes a lethal molting disruption in Daphnia pulicaria. Appl Environ Microbiol. 2004;70:5047–5050. doi: 10.1128/AEM.70.8.5047-5050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Zheng T, Bruner SD. Structural basis for phosphopantetheinyl carrier domain interactions in the terminal module of nonribosomal peptide synthetases. Chem Biol. 2011;18:1482–1488. doi: 10.1016/j.chembiol.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koehnke J, et al. The mechanism of patellamide macrocyclization revealed by the characterization of the PatG macrocyclase domain. Nat Struct Mol Biol. 2012;19:767–772. doi: 10.1038/nsmb.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang B, Zhao A, Novick RP, Muir TW. Key driving forces in the biosynthesis of autoinducing peptides required for staphylococcal virulence. Proc Natl Acad Sci USA. 2015;112:10679–10684. doi: 10.1073/pnas.1506030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan SJ, Rajniak J, Cheung WL, Link AJ. Construction of a single polypeptide that matures and exports the lasso peptide microcin J25. ChemBioChem. 2012;13:367–370. doi: 10.1002/cbic.201100596. [DOI] [PubMed] [Google Scholar]
- 27.Oman TJ, Knerr PJ, Bindman NA, Velásquez JE, van der Donk WA. An engineered lantibiotic synthetase that does not require a leader peptide on its substrate. J Am Chem Soc. 2012;134:6952–6955. doi: 10.1021/ja3017297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koehnke J, et al. Structural analysis of leader peptide binding enables leader-free cyanobactin processing. Nat Chem Biol. 2015;11:558–563. doi: 10.1038/nchembio.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortega Ma, et al. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature. 2014;517:509–512. doi: 10.1038/nature13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burkhart BJ, Hudson GA, Dunbar KL, Mitchell DA. A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat Chem Biol. 2015;11:564–570. doi: 10.1038/nchembio.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong SH, et al. The enterococcal cytolysin synthetase has an unanticipated lipid kinase fold. Elife. 2015;4:e07607. doi: 10.7554/eLife.07607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie L, et al. Lacticin 481: In vitro reconstitution of lantibiotic synthetase activity. Science. 2004;303:679–681. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
- 33.Cheung WL, Pan SJ, Link AJ. Much of the microcin J25 leader peptide is dispensable. J Am Chem Soc. 2010;132:2514–2515. doi: 10.1021/ja910191u. [DOI] [PubMed] [Google Scholar]
- 34.Philmus B, Guerrette JP, Hemscheidt TK. Substrate specificity and scope of MvdD, a GRASP-like ligase from the microviridin biosynthetic gene cluster. ACS Chem Biol. 2009;4:429–434. doi: 10.1021/cb900088r. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt EW, et al. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller S, et al. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal Chem. 2012;84:5066–5073. doi: 10.1021/ac3007522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabsch W. XDS. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD. Substructure search procedures for macromolecular structures. Acta Crystallogr Sect D Biol Crystallogr. 2003:1966–1973. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr Sect D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldenberg O, Erez E, Nimrod G, Ben-Tal N. The ConSurf-DB: Pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 2009;37:323–327. doi: 10.1093/nar/gkn822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dolinsky TJ, et al. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:522–525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris GM, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2790. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.