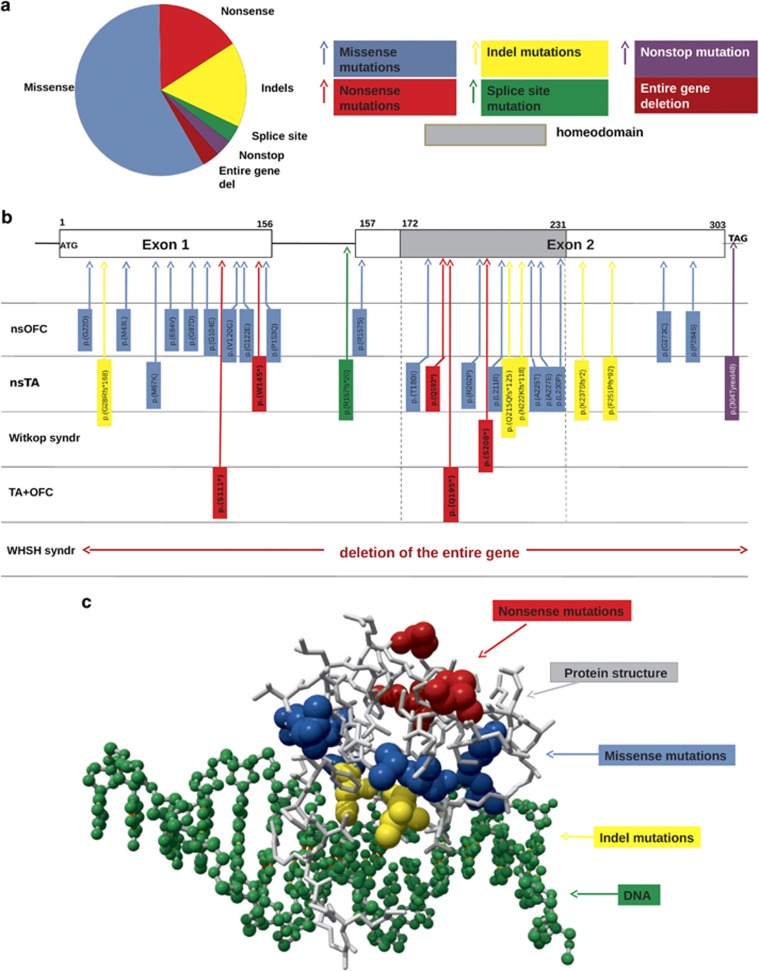
Figure 1.
Distribution of all MSX1 mutations included in this review. Reference sequence NC_000004.12, chrom 4, GRCh38.p2 (GCF_000001405.28); NP 002439.2; NM 002448,3 was used on which the variant description of nucleotides and amino acids are based. (a) Pie diagram with the ‘in-frame' (blue; n=18) versus 13 ‘truncating' mutations of which 5 nonsense mutations (red), 5 indel mutations (yellow), 1 splice site mutation (green), 1 nonstop mutation (purple), as well as 1 whole gene deletion (brown). The in-frame mutations were only missense mutations. (b) Mapping of the MSX1 mutations with associated disease phenotypes to coding and non-coding structures of MSX1. The horizontal boxes on top represent exon 1 and exon 2; the line between the two exons represents the intron; the grey box is the homeodomain (HD) coding area; ATG, start codon; TAG, stop codon. The mutant MSX1 variants in the vertical blue boxes represent the in-frame missense variants; those in the red boxes the nonsense mutations, in the yellow boxes the indel mutations, in the green box the splice site mutation and in the purple box the nonstop mutation. (c) In the yet experimentally determined structural part of the MSX1 protein (PSI; Protein Model Portal; 1ig7; Msx-1 Homeodomain/DNA Complex Structure; residues 173-230; http://www.proteinmodelportal.org/query/uniprot/P28360) the mutations are indicated in the same colors as in parts (a) and (b) of Figure 1: blue for the missense mutations; red for the nonsense mutations and yellow for the indel mutations; DNA is depicted in green; and protein structure in grey. From the location in the MSX1 structure, the indel mutations can be predicted to severely disturb the DNA binding. Although the same holds true for the missense mutations, they will disturb the DNA binding with variable severity. The red residues become stop codons and thus represent the first absent residues in these mutations; their effect cannot be predicted. A full color version of this figure is available at the European Journal of Human Genetics journal online.