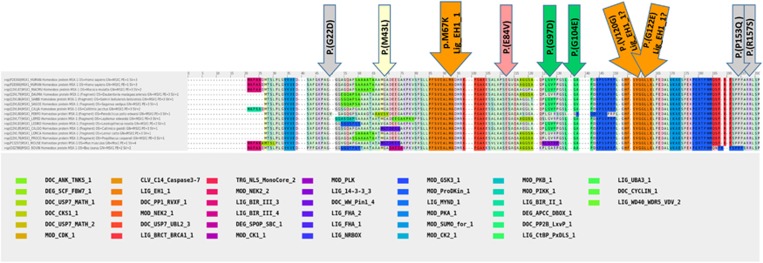
Figure 2.
Panel with the amino acid sequences and motifs in the natively unfolded part of the MSX1 protein According to the Knowledge based Multiple sequence Alignment for intrinsically Disordered proteins (KMAD; DOI:10.1093/bioinformatics/btv663; Lange et al, 2016) most of the prediction programs attribute the term ‘natively unfolded') to this part of the MSX1 protein lying N-terminally to the homeodomain. Although many of the sequence motifs of this natively unfolded part of the MSX1 protein have not yet been experimentally associated to a function (KMAD-align; DOI:10.1093/bioinformatics/btv663), an important role is obvious when the amino-acid (AA) sequence is highly conserved. This is the case for the AA sequence in the large orange block under the orange arrow which refers to the LIG_EH1_1 motif where the M67K mutation causing ns TA is residing. This mutation will dramatically disrupt the function of this LIG_EH1_1 motif conserved sequence. All other arrows point to the MSX1 variants which are all associated with ns OFC. It can be noticed that the motifs/sequences containing these variants show a lesser degree of conservation. MOD: stands for variants found in mouse data. Both MOD_SUMO and MOD_CDK1 (see color legend), have previously been mentioned in relation to cleft lip and palate development in mice. A full color version of this figure is available at the European Journal of Human Genetics journal online.