Abstract
In approximately 20% of individuals with Kagami-Ogata syndrome (KOS14, MIM 608149), characterized by a bell-shaped thorax with coat-hanger configuration of the ribs, joint contractures, abdominal wall defects and polyhydramnios during the pregnancy, the syndrome is caused by a maternal deletion of the imprinted gene cluster in chromosome 14q32.2. Most deletions reported so far included one or both of the differentially methylated regions (DMRs) – DLK1/MEG3 IG-DMR and MEG3-DMR. We present two unrelated families with two affected siblings each, presenting with classical KOS14 due to maternally inherited microdeletions. Interestingly, all four patients have lived through to adulthood, even though mortality rates for patients with KOS14 due to a microdeletion are relatively high. In the first family, none of the DMRs is included in the deletion and the methylation status is identical to that of controls. Deletions that do not encompass the DMRs in this region are thus sufficient to elicit the full KOS14 phenotype. In the second family, a partially overlapping deletion including both DMRs and MEG3 was detected. In summary, we show that patients with KOS14 can live into adulthood, that causal deletions do not have to include the DMRs and that consequently a normal methylation pattern does not exclude KOS14.
INTRODUCTION
The human chromosome region 14q32 contains a number of imprinted genes that are either expressed from the paternal (eg, DLK1 and RTL1) or from the maternal allele (eg, MEG3, RTL1as and MEG8). The locus contains two differentially methylated regions (DMRs) involved in imprinting regulation, which are unmethylated on the maternal allele. The germline-derived DLK1-MEG3 intergenic DMR (IG-DMR) is located between the DLK1 and MEG3 genes and operates as an imprinting control center in the placenta, whereas the post-fertilization-derived MEG3-DMR resides in the promoter region of the MEG3 gene and functions as an imprinting control center in the embryo.1, 2, 3 The IG-DMR regulates the methylation status of the MEG3-DMR.4, 5 A maternal deletion of the IG-DMR therefore affects the methylation status of the MEG3-DMR, while a maternal deletion of the MEG3-DMR does not affect the methylation status of the IG-DMR.
Paternal uniparental disomy of chromosome 14, or UPD(14)pat, involving the imprinted region on 14q32.2 is known as Kagami-Ogata syndrome (KOS14, MIM #608149). Patients are characterized by a small bell-shaped thorax with a coat-hanger configuration of the ribs, abdominal wall defects, joint contractures and polyhydramnios during the pregnancy.6, 7 The reciprocal disorder (UPD(14)mat), nowadays known as Temple syndrome (TS14, MIM #616222) is characterized by short stature, developmental delay and early puberty.6 KOS14 is caused by UPD(14)pat in approximately 65% of cases, by maternal microdeletions in the DLK1/MEG3 region in 19% of cases and by hypermethylation of the DMRs in 15% of patients.8 It has been suggested that an excessive RTL1 expression caused by absence of the antisense transcript RTL1as and absent MEG expression underlie many of the phenotypic consequences of the KOS14, but the precise genotype-phenotype correlation has not been fully elucidated as yet.9
Here, we present two families, both with two affected siblings, with a maternal microdeletion on chromosome 14q32.2. The first family carries a deletion that does not include any of the two DMRs and does not show methylation abnormalities. The second family carries a deletion that contains both DMRs and part of the MEG3 gene, but not the RTL1as gene.
MATERIALS AND METHODS
Subjects
Two families (family A and family B), both with two affected siblings, were referred for microarray analysis to search for a cause of their disease. All patients displayed the clinical features of Kagami-Ogata syndrome during childhood, which was not yet described in literature at that time and thus not diagnosed. Extensive clinical descriptions of all patients, including pedigrees, clinical pictures and radiographical thorax images are available in the Supplementary Information.
Microarray
DNA was extracted from peripheral blood using standard methods. SNP array analysis in patients AII.1 and AII.2 was performed using a HumanCytoSNP-12 v2.1 beadchip on an iScan system, following standard protocols as provided by the manufacturer (Illumina, San Diego, CA, USA). CNV analysis was performed using CNV-Webstore.10 Familial relationships were validated based on the SNP pattern inheritance. In patients BII.1 and BII.3, array analysis was performed using the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA). The data of all patients are deposited in the Decipher database with submission IDs 326793 (AII.1), 326787 (A.II.2), 327656 (BII.1) and 327657 (BII.3) (https://decipher.sanger.ac.uk/index).
Methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA)
Gene dosage of the imprinted DLK1/MEG3 locus and methylation of the MEG3 promoter region on chromosome 14q32 were analyzed by MS-MLPA using the SALSA MLPA KIT ME032-A1 kit (MRC Holland, Amsterdam, the Netherlands). Hybridization, ligation and PCR reactions were performed according to the manufacturer's instructions. Amplification products were analyzed by capillary electrophoresis using the ABI3100 capillar sequencer (Applied Biosystems Inc., Foster City, CA, USA). Data analysis was carried out using the Gene Marker Software (Softgenetics, State College, PA, USA).
DNA methylation analysis of the DLK1-MEG3 IG-DMR by next-generation bisulfite sequencing
Bisulfite treatment was performed using the EZ DNA Methylation-Gold Kit (Zymo Research Europe, Freiberg, Germany) according to the manufacturer's instructions. For each individual, bisulfite amplicon libraries were generated and sample-specific barcode sequences were added. The amplicons were purified, diluted and clonally amplified in an emulsion PCR before sequencing on the Roche/454 GS junior system (Branford, CT, USA). A detailed description has been published previously.5, 11 For subsequent data analysis, the Amplikyzer software was used (https://bitbucket.org/svenrahmann/amplikyzer/downloads). A minimum of 1200 reads for each sample was obtained. Only reads with a conversion rate over 95% were considered.
Copy number profiling using qPCR
Copy number profiling using quantitative PCR was performed as described by D'haene et al.12 In short, several primer pairs were designed to amplify 80–150 basepairs each. Quantitative PCR on patient DNA samples was performed using the qPCR Mastermix Plus for SYBR Green I – no ROX (Eurogentec, Liège, Belgium) according to the manufacturer's protocol, on a Lightcycler480 (Roche, Basel, Switzerland). Data analysis was performed using qbase+ (Biogazelle, Ghent, Belgium).
Targeted locus amplification sequencing (TLA sequencing)
Viably frozen cells from an EBV-cell line of patient AII.1 were sent to Cergentis B.V. (Utrecht, the Netherlands) for TLA sequencing. Sample preparation, sequencing and data analysis were performed as described by de Vree et al.13
Sanger sequencing
Primers were designed using Primer3Plus.14 PCR was performed with the GoTaq G2 Polymerase kit (Promega Benelux, Leiden, the Netherlands) according to the guidelines of the manufacturer. PCR cleanup was performed using alkaline phosphatase (Roche, Basel, Switzerland) and exonuclease I (Bioké, Leiden, the Netherlands). Sanger sequencing reactions were performed using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit with readout on an ABI3130XL sequencer (Applied Biosystems Inc.). CLC DNA Workbench 5.0.2 (CLC bio, Aarhus, Denmark) was used for data analysis.
RESULTS
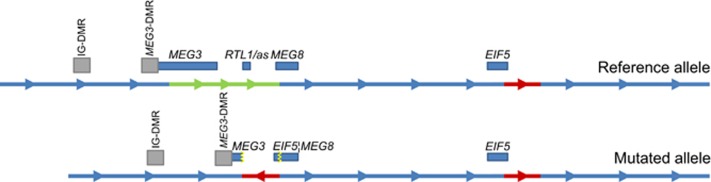
The phenotypes of the patients in family A (AII.1 and AII.2) strongly suggested a diagnosis of UPD(14)pat when first evaluated in the clinic in the early 1990s. Karyotypes were performed to search for a Robertsonian translocation, but no abnormalities were detected. Microsatellite analysis, the most sensitive detection method at that time, did not detect any evidence for UPD. Further genetic testing was not performed until recently, when SNP array analysis became available and revealed a microdeletion in both siblings on chromosome 14q32.2 within the imprinted domain of this chromosomal region (hg19 chr14:g.(101290183_101297515)_(101361403_101367310)del). Microarray analysis of the mother (AI.2) was suggestive for the presence of this microdeletion in a mosaic state (Supplementary Information). To further delineate the critical region for the KOS14 phenotype in this family, we performed copy number profiling using quantitative polymerase chain reactions (qPCR). Using these qPCR assays, we refined the breakpoints to a region of minimally 65.8 kb and maximally 66.5 kb. Copy number qPCR confirmed the presence of the exact same deletion in the mother in mosaic state. Attempts to span the breakpoint by PCR assays failed to produce an amplicon, suggesting the presence of a more complex rearrangement. Targeted locus amplification (TLA) sequencing was performed in patient AII.1 and revealed that the deletion contained a 16 kb insertion, including a part of the EIF5 gene derived from a downstream chromosomal region. Copy number qPCR assays specifically targeting this sequence showed that three copies of the insertion were present in patients AII.1 and AII.2 and confirmed the mosaic state in the mother. A schematic overview of the rearrangement, depicting that the duplicated sequence was inserted in reverse orientation is shown in Figure 1. Sanger sequencing of the fusion points confirmed the exact rearrangement in patient AII.1 and showed that the same rearrangement is present in patient AII.2 and the mother of these siblings (hg19 chr14:g.(101297067)_(101362959)del, hg19 chr14:g.(103809080)_(103825690)inv_ins). The deletion affects three maternally expressed genes: all exons except for the first three of the maternally expressed gene 3 (MEG3), the entire RTL1as gene and the upstream region and first exon of MEG8. None of the two DMRs that reside upstream of the MEG3 gene (IG-DMR and MEG3-DMR) were involved in the deleted region. Methylation analysis by MS-MLPA and next-generation bisulfite sequencing revealed a normal methylation pattern at the MEG3-DMR and the IG-DMR loci (Figure 2, Supplementary Information).
Figure 1.
Schematic representation of the causal structural rearrangement in family A. Upper scheme represents the normal state and lower scheme represents the rearranged state of the 14q region in the patients. The genes involved in the rearrangement are represented by the blue boxes; gray boxes indicate a DMR. Arrowheads indicate the 5′ to 3′ direction. The green section is deleted in the patients, the red section is duplicated and inserted in between the deletion breakpoints in reverse orientation. A full color version of this figure is available at the European Journal of Human Genetics journal online.
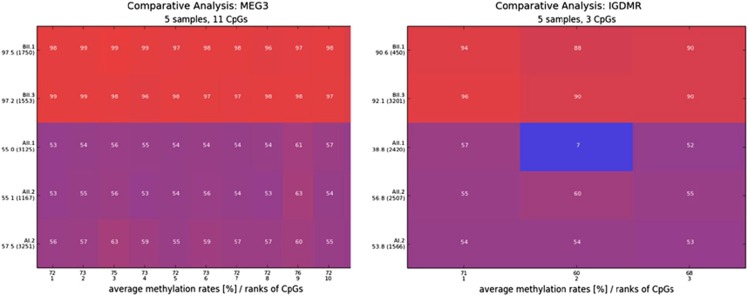
Figure 2.
Comparative methylation heatmap obtained by next-generation bisulfite sequencing of MEG3 and the DLK1/MEG3 IG-DMR. Patient BII.1 and BII.3 showed hypermethylation due to the deletion of the unmethylated maternal allele at both loci, whereas patients AII.1 and AII.2 and their mother showed normal methylation values around 50%. Patient AII.1 showed hypomethylation at CpG2 of the IG-DMR because she is heterozygous or homozygous for a C to T exchange (rs1458662425:C>T (hg19. chr14:g.101277356C>T)) on the methylated paternal allele. Each square represents a CpG dinucleotide with its average methylation level and each line a specific sample. The average methylation over the analyzed region is given in percentage under the individual sample together with the number of analyzed reads in brackets. The methylation was analyzed over 11 CpGs at the MEG3-DMR and 3 CpGs at the DLK1/MEG3 IG-DMR. A full color version of this figure is available at the European Journal of Human Genetics journal online.
Although patients BII.1 and BII.3 presented with typical clinical findings of Kagami-Ogata syndrome, the syndrome was not yet described in literature at the time they were born. Thus, a diagnosis was not established until their healthy brother came to the clinic asking for genetic counseling because of two affected siblings with intellectual disability. Conventional chromosome analysis in the family showed no signs of a chromosomal rearrangement. However, SNP array analysis identified a 130 kb microdeletion in 14q32 hg19 chr14:g.(101204495_101209293)_(101340188_101340732)del in patient BII.1. The deletion includes MEG3 as well as both imprinting control regions MEG3-DMR and IG-DMR. Presence of the deletion in patient BII.1 was confirmed by qPCR and MLPA. Dosage analysis by qPCR and MLPA in the affected brother (BII.3) showed that he is also a carrier. The deletion was absent in the unaffected brother and the parents. Methylation analysis by MS-MLPA of the MEG3-DMR showed hypermethylation, consistent with paternal inheritance of the remaining allele and thus indicating that the deletion is maternally inherited. MS-MLPA revealed the same hypermethylation in the affected brother (patient BII.3) but normal methylation patterns in the parents (Figure 2, Supplementary Information). As the mother does not show any evidence for the deletion in peripheral leukocytes, MS-MLPA was additionally performed on DNA extracted from epithelium of the oral mucosa to identify a potential somatic mosaicism. Again, dosage and methylation analysis did not reveal any evidence of a deletion. It is therefore plausible that she has a germline mosaicism for this deletion, although a very low-grade somatic mosaicism not detectable by MS-MLPA cannot be excluded. In line with this hypothesis, microsatellite analysis of the 14q32 region using six informative markers (D14S987, D14S979, D14S267, D14S250, D14S1006 and D14S1010) showed that both affected siblings and the healthy brother have inherited the same maternal allele. However, for the marker D14S985, which lies inside the deletion (in intron 3 of MEG3), the healthy brother has inherited both a paternal and maternal allele, whereas his affected siblings lack a maternal allele owing to the deletion.
DISCUSSION
A detailed description of the clinical features in combination with the molecular causes of disease is helpful in correlating the genotype with the phenotype and in elucidating the specific role of the individual transcripts in the 14q32.2 imprinted region in the emergence of the KOS14 phenotype.15 Here, we add two pairs of siblings from two unrelated families with a maternally inherited microdeletion in the 14q32 imprinting region to the cohort of 34 recently reported KOS14 patients.15 All four affected individuals survived into adulthood, with relatively mild phenotypes compared with the patients with KOS14 due to microdeletions that have been described.15 Mortality rates of KOS14 appear to be associated with premature birth and mutation type, with death typically occurring before the age of four. In a cohort of 34 patients described by Kagami et al.,15 the survival rate of deletion carriers was only 50%, while a survival rate of 78% was observed for the UPD(14)pat group and even 100% for the individuals with epimutations. Causes of death were variable, but overall respiratory problems seem to have a major role.
Patient AII.1 presented the typical coat-hanger sign of the ribs and the bell-shaped deformity of the thorax that is characteristic for KOS14. The deformity diminished during development and to date she is an ambitious young lady, who suffers from recurrent respiratory infections and easy tiring. Her brother, patient AII.2, not only showed the typical deformities of the thoracic cage as classical KOS14, but also presented with abdominal wall defects, distal arthrogryposis deformities and polyhydramnios during the pregnancy, features commonly observed in KOS14 patients. To date, he suffers from mild intellectual disability and persistent hypotonia. Even though patients AII.1 and AII.2 carry the exact same deletion, she is less severely affected than her younger brother. The intellectual disability seen in patient AII.2 is a feature commonly seen in KOS14 and thus might be intrinsic to the disease, but in his case, might also be the result of unnoticed hypoxic periodes in the neonatal period due to the skeletal malformations of the thorax. In contrast, his sister AII.2 finished academic education and the KOS14 phenotypic features she presented with during early childhood almost completely normalized. In a previously reported sibling pair carrying a 5.8 kb deletion affecting the MEG3-DMR and the first three exons of the MEG3 gene, the clinical course of the girl was also in general milder than that of her older brother.5 Even though this may well be a coincidental observation, gender-determined factors cannot be ruled out at the moment.
Patient BII.1 was born prematurely and showed a bell-shaped thorax, general muscular hypotonia, bilateral inguinal hernia and diastasis recti. He displayed mild developmental delay, but managed to complete vocational training and works as an office clerk to date. His younger brother, patient BII.3, also showed the bell-shaped deformity of the thorax, combined with respiratory distress, bilateral inguinal hernia and general muscular hypotonia. He completed special education and currently works in a sheltered workshop. The patients we present here display (mild) clinical features due to the maternally inherited deletion on chromosome 14, but live through adulthood which is remarkable for patients with Kagami-Ogata syndrome caused by microdeletions. All of the patients, except patient AII.2, were able to finish education to some extent and obtain a job that is consistent with their respective education levels. Overall, a reduction in the thoracic abnormalities was observed during the development, suggesting that these are deformations rather than malformations. These skeletal malformations might be attributed to reduced bone hardness, through which the deformation of the skull seen in patient AII.2 could also be explained. As these skeletal deformations, especially the narrow thorax, can be detected prenatally by ultrasound, intensive neonatal care should be provided as soon as possible upon birth to prevent hypoxia-induced damage.
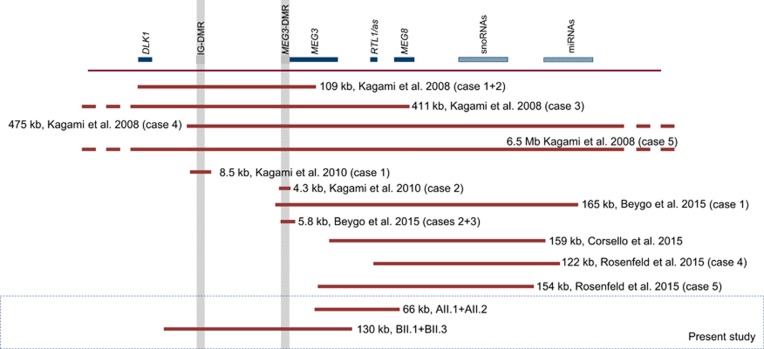
To our knowledge, 13 patients with a UPD(14)pat phenotype caused by a maternal 14q32 deletion have been reported.3, 4, 5, 16, 17 The precise localization of these is depicted in Figure 3. The clinical characteristics of all deletion carriers, including the cases described here are summarized in Table 1. Kagami et al.3 reported four different deletions in five cases. The phenotype ranged from typical (eg, cases 1+2) to relatively mild KOS14 (eg, cases 3–5). Subsequently, two patients with a severe KOS14 phenotype were described, both carrying a remarkably small deletion that only encompasses one of the two DMRs.3, 4 Beygo et al.5 described two novel maternal microdeletions as the cause of the KOS14 phenotype in three additional cases, including a sibling pair. All initial 10 cases had a deletion that included at least one or both imprinting control centers (IG-DMR and MEG3-DMR). Because of the hierarchical regulation of these imprinting clusters, in which the IG-DMR influences the methylation of the MEG3-DMR, aberrant methylation of the MEG3-DMR was likely present in all. However, the deletion detected in our family A does not include either one of the DMRs, suggesting that the involvement of the DMRs is not essential for the development of the disorder. Using MS-MLPA and bisulfite sequencing, abnormal methylation was ruled out in family A, indicating that the maternally inherited absence of MEG3, RTL1as and MEG8 per se is causative for the phenotype in this family rather than aberrant methylation of the DMRs. Interestingly, TLA sequencing unraveled the presence of a complex rearrangement, in which a 16 kb sequence, derived from a region downstream of the deletion, was duplicated and inserted in reverse orientation. The precise molecular mechanism through which this rearrangement arose is unclear, but it is concordant with a Fork Stalling and Template Switching (FoSTes) model.18, 19 As the inserted sequence does not contain the complete EIF5 gene and two intact copies-one on each allele-of this gene are still present, we consider it unlikely that the EIF5 gene contributes to the clinical presentation of KOS14 in our patients.
Figure 3.
Reported microdeletions in 14q32.2 leading to the KOS14 phenotype. Horizontal bars represent all cases known to date carrying a microdeletion on their maternal allele, the length of the bars indicates the size and genes encompassed by the respective deletions. Gray shaded boxes reflect the position of DMRs. A full color version of this figure is available at the European Journal of Human Genetics journal online.
Table 1. Clinical characteristics of patients with Kagami-Ogata Syndrome caused by a maternal microdeletion in chr14q32.2.
| Kagami et al.3 | Kagami et al.4 | Beygo et al.5 | Corsello et al.16 | Rosenfeld et al.17 |
Current study |
||||
|---|---|---|---|---|---|---|---|---|---|
| AII.1 | AII.2 | BII.1 | BII.3 | ||||||
| Number of patients | 5 | 2 | 3 | 1 | 2 | ||||
| Gender (Female:Male) | 3:2 | 2:0 | 1:2 | Female | 2:0 | Female | Male | Male | Male |
| Molecular findings | |||||||||
| IG-DMR of maternal origin | Deleted (5/5) | Deleted (1/2) | Unaffected | Unaffected | Unaffected | Unaffected | Unaffected | Deleted | Deleted |
| MEG3-DMR of maternal origin | Deleted (5/5) | Deleted (1/2) | Deleted (3/3) | Unaffected | Unaffected | Unaffected | Unaffected | Deleted | Deleted |
| Prenatal | |||||||||
| Polyhydramnios | + (4/5) | + (1/2) | + (3/3) | + | + (2/2) | ? | + | + | + |
| Other | Ultrasound finding of bell-shaped, narrow thoracic deformity | Premature labor from week 28 onward | Sonographically suspected encephalocele (not confirmed postnatally) | ||||||
| Neonatal | |||||||||
| Gestational age in weeks (range) | 27–35 | 28–33 | 32–35 | 35 | ? | 41 | 35 | 32 | 35 |
| Hypotonia | ? | ? | + (3/3) | ? | + (1/2) | + | + | + | + |
| Feeding difficulties | ? | ? | + (2/3) | + | ? | − | + | − | + |
| Thorax | |||||||||
| Bell-shaped thorax with coat-hanger ribs | + (5/5) | + (2/2) | + (3/3) | + | + (2/2) | + | + | + | + |
| Mechanicalventilation | + (5/5) | + (2/2) | + (3/3) | + | ? | − | + | − | + |
| Other | Pectus excavatum | Pectus excavatum | |||||||
| Abdomen | |||||||||
| Diastasis recti | + (2/5) | ? | + (1/3) | + | − | + | − | ||
| Omphalocele | − | + (2/2) | ? | − | − | − | − | ||
| Other | Bilateral hydronephrosis (1/2) | Umbilical hernia (1/3), inguinal hernia (1/3) | Bilateral inguinal hernia | Bilateral inguinal hernia and umbilical hernia | |||||
| Extremities | |||||||||
| Joint contractures | + (2/5) | + (1/2) | ? | + | − | − | + | − | − |
| Hyperextensibility of small joints | ? | ? | − | ? | ? | + | |||
| Development | |||||||||
| Education | ? | ? | ? | ? | ? | ++ | − | +/− | +/− |
| Developmental delay | + (3/5) | ? | + (2/3) | ? | ? | − | + | +/− | + |
Abbreviation: IG-DMR, intergenic differentially methylated region. +, feature is present; −, feature is absent; ?, feature is not described as present or absent.
While preparing this manuscript, three additional microdeletions were reported that did not include the DMRs, like in our family A.16, 17 As visualized in Figure 3, there is no smallest region of overlap common to all 13 microdeletions. It is therefore likely that several regulatory mechanisms can be disrupted at different positions within the 14q32.2 chromosomal region with a similar clinical outcome. It is generally accepted that aberrant expression of the maternally transcribed copies of MEG3, MEG8 or RTL1as – either through the disruption of the imprinting control centers IG-DMR and MEG3-DMR or through direct disruption of the genes by a deletion – has a crucial role in the pathogenesis of KOS14, although an effect of other regulatory sequences cannot be excluded. Little is known about the function of the long non-coding RNAs – MEG3 and MEG8. Maternal RTL1as transcripts function to repress RTL1 transcripts from the paternal allele and absence of RTL1as leads to an increase in RTL1 expression.20 Increased expression of RTL1, potentially combined with the effects of absent expression of MEGs has been hypothesized to have a key role in the pathogenesis of KOS14.9 This hypothesis is strengthened by the observation that the RTL1as transcript is commonly deleted in the microdeletions that do not affect the imprinting control centers. The deletion in family B contains both imprinting control centers and MEG3, but not the RTL1as transcript, neither MEG8. However, it is not known whether in our patients BII.1 and BII.3, the transcription of RTL1as and MEG8 is also affected. In a mouse model, absence of the first exons of Gtl2, the murine homolog of MEG3, has been reported to silence the expression of all maternally expressed genes.21
In summary, we show that some patients with KOS14 can live into adulthood, which should have an impact on handling these patients in a neonatal care unit. In addition, we demonstrate that deletions not including the IG-DMR nor the MEG3-DMR and devoid of methylation abnormalities can lead to KOS14, emphasizing the importance of the non-coding transcripts in the region.
Acknowledgments
We thank Dr Max van Min for his advice on TLA sequencing, Christina Lich for technical assistance and Jasmin Beygo for her help with figures and helpful discussions. IMvdW is funded by the Special Research Fund of the University of Antwerp (Bijzonder Onderzoeksfonds (BOF-IWT)). GV is a postdoctoral fellow of the Research Fund Flanders (FWO). Part of the work was funded by the Bundesministerium für Bildung und Forschung (Network Imprinting diseases, 01GM1513A).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Lin SP, Youngson N, Takada S et al: Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet 2003; 35: 97–102. [DOI] [PubMed] [Google Scholar]
- Paulsen M, Takada S, Youngson NA et al: Comparative sequence analysis of the imprinted Dlk1-Gtl2 locus in three mammalian species reveals highly conserved genomic elements and refines comparison with the Igf2-H19 region. Genome Res 2001; 11: 2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami M, Sekita Y, Nishimura G et al: Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet 2008; 40: 237–242. [DOI] [PubMed] [Google Scholar]
- Kagami M, O'Sullivan MJ, Green AJ et al: The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet 2010; 6: e1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beygo J, Elbracht M, de Groot K et al: Novel deletions affecting the MEG3-DMR provide further evidence for a hierarchical regulation of imprinting in 14q32. Eur J Hum Genet 2015; 23: 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton VR, Shaffer LG: Search for imprinted regions on chromosome 14: comparison of maternal and paternal UPD cases with cases of chromosome 14 deletion. Am J Med Genet 2000; 93: 381–387. [DOI] [PubMed] [Google Scholar]
- Kagami M, Nishimura G, Okuyama T et al: Segmental and full paternal isodisomy for chromosome 14 in three patients: narrowing the critical region and implication for the clinical features. Am J Med Genet A 2005; 138A: 127–132. [DOI] [PubMed] [Google Scholar]
- Kagami M, Kato F, Matsubara K, Sato T, Nishimura G, Ogata T: Relative frequency of underlying genetic causes for the development of UPD(14)pat-like phenotype. Eur J Hum Genet 2012; 20: 928–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata T, Kagami M: Kagami-Ogata syndrome: a clinically recognizable upd(14)pat and related disorder affecting the chromosome 14q32.2 imprinted region. J Hum Genet 2015; 61: 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeweyer G, Reyniers E, Wuyts W, Rooms L, Kooy RF: CNV-WebStore: online CNV analysis, storage and interpretation. BMC Bioinformatics 2011; 12: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beygo J, Citro V, Sparago A et al: The molecular function and clinical phenotype of partial deletions of the IGF2/H19 imprinting control region depends on the spatial arrangement of the remaining CTCF-binding sites. Hum Molr Genet 2013; 22: 544–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Haene B, Vandesompele J, Hellemans J: Accurate and objective copy number profiling using real-time quantitative PCR. Methods 2010; 50: 262–270. [DOI] [PubMed] [Google Scholar]
- de Vree PJ, de Wit E, Yilmaz M et al: Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat Biotechnol 2014; 32: 1019–1025. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA: Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 2007; 35: W71–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami M, Kurosawa K, Miyazaki O, Ishino F, Matsuoka K, Ogata T: Comprehensive clinical studies in 34 patients with molecularly defined UPD(14)pat and related conditions (Kagami-Ogata syndrome). Eur J Hum Genet 2015; 23: 1488–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsello G, Salzano E, Vecchio D et al: Paternal uniparental disomy chromosome 14-like syndrome due a maternal de novo 160kb deletion at the 14q32.2 region not encompassing the IG- and the MEG3-DMRs: Patient report and genotype-phenotype correlation. Am J Med Genet A 2015; 167A: 3130–3138. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JA, Fox JE, Descartes M et al: Clinical features associated with copy number variations of the 14q32 imprinted gene cluster. Am J Med Genet A 2015; 167A: 345–353. [DOI] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR: A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 2007; 131: 1235–1247. [DOI] [PubMed] [Google Scholar]
- Liu P, Carvalho CM, Hastings P, Lupski JR: Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev 2012; 22: 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami M, Matsuoka K, Nagai T et al: Paternal uniparental disomy 14 and related disorders: placental gene expression analyses and histological examinations. Epigenetics 2012; 7: 1142–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetatos L, Hatzimichael E, Londin E et al: The microRNAs within the DLK1-DIO3 genomic region: involvement in disease pathogenesis. Cell Mol Life Sci 2013; 70: 795–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.