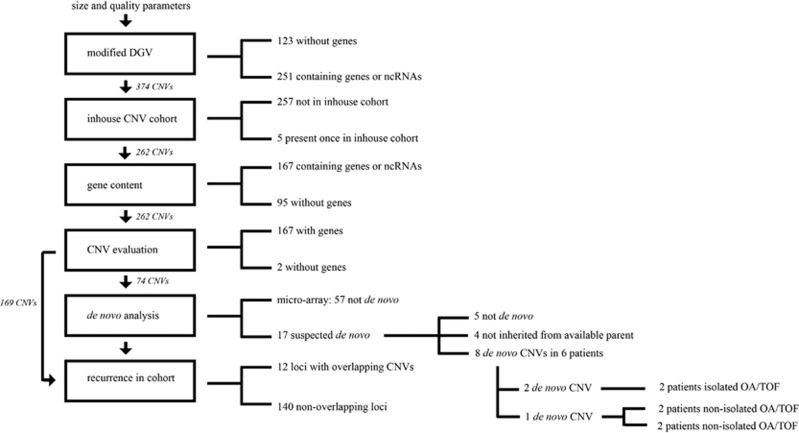
Figure 1.
Filtering and prioritizing CNVs. After quality control and manual evaluation of CNVs, 374 CNVs larger than 30 kb, either absent or rare in the modified Database of Genomic Variants incorporated in the Nexus software, remained. Out of 374, 123 did not contain genes. In all, 257 were absent and 5 were present once in our in-house control database. These 262 CNVs were either gene-rich – containing genes – (n=167) or gene-poor (n=95). Two gene-poor CNVs were suspected of being de novo in microarray trio analysis. Eight out of 74 evaluated CNVs were de novo. Almost all of the rare CNVs (140) were widely distributed over the genome. However, our analysis yielded a total of 12 loci – containing 29 CNVs – which were affected by a rare CNV more than once and were present in more than one patient.