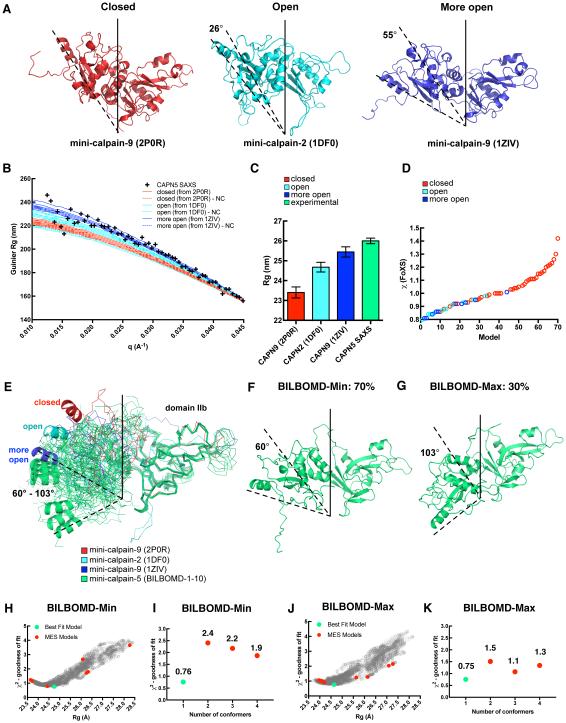
Fig. 4. Fit of different mini-calpain-5 models to SAXS data.
(A) Calpain protease core structures adopt one of three conformations: closed (mini-calpain-9; PDB ID 2P0R), open (mini-calpain-2; PDB ID 1DFO), and more open (mini-calpain-9; PDB ID 1ZIV). (B) Low q (Guinier) region and fit of representative rigid homology models of closed, open and more open forms of mini-calpain-5 to SAXS data. The homology models with the floppy N- and C-terminus trimmed are labeled as “-NC”. (C) Rg calculated for the representative homology models of mini-calpain-5 compared with the Guinier Rg of SAXS data. (D) Fits of 70-homology models based off 4 closed and 3 open forms to SAXS data. (E) The 10 best-fit BILBOMD models of mini-calpain-5 starting with a homology model based off 1ZIV are shown in green. DIIb of all the structures have been superimposed to highlight the relative orientation of DIIa. Rotation angles calculated using DynDom with respect to mini-calpain-1 (2ARY) are reported. The BILBOMD models fell into two clusters, represented by BILBOMD-Min (F) and BILBOMD-Max (G) that comprised 70% and 30% of the best-fit conformations, respectively. (H) Graph representing the goodness of fit (χ2) for 4,000 conformations generated for BILBOMD-Min with their Rg values. The green and red dots represent the best-fit model and the minimal ensemble search (MES) models that were weighted, respectively. (I) MES fits for BILBOMD-Min to SAXS data indicate that the single conformer has the lowest χ2 and is the best-fit model. (J) Goodness of fit (χ2) for 4,000 conformations generated for BILBOMD-Max with their Rg values. (K) MES fits for BILBOMD-Max to SAXS data.