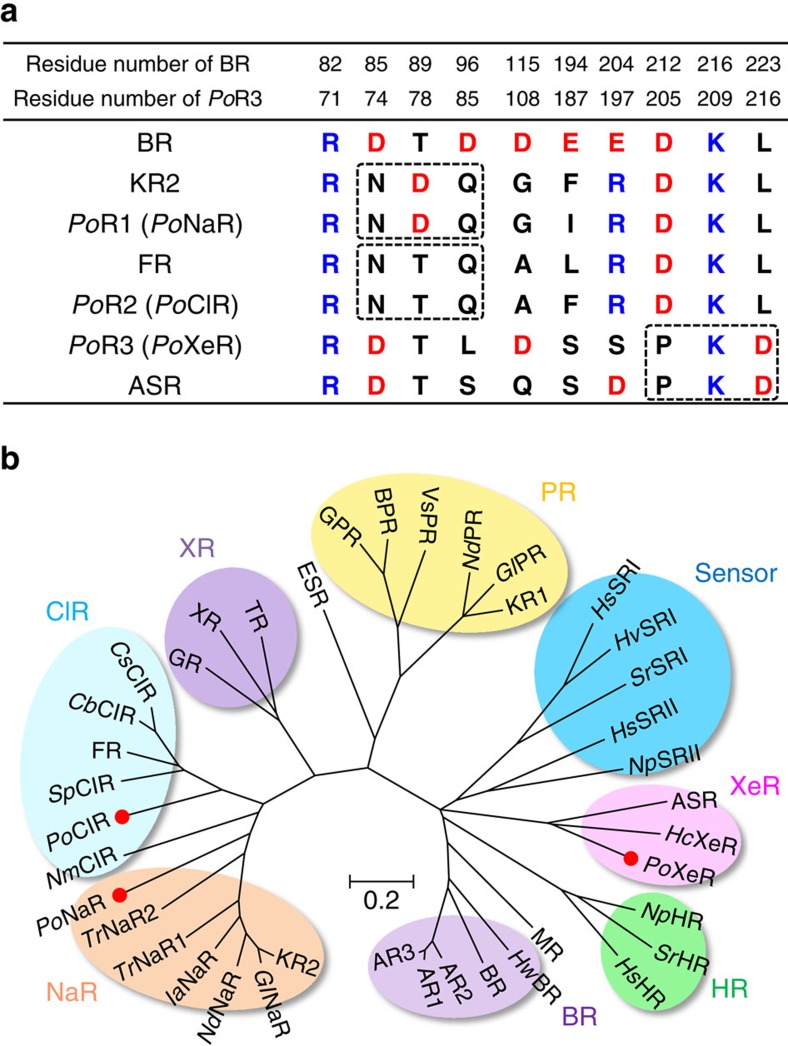
Figure 2. Amino-acid sequence alignment of important residues and phylogenetic tree of microbial rhodopsins.
(a) Important residues for the function of microbial rhodopsins. BR, KR2, FR and ASR are outward H+ pump, outward Na+ pump, inward Cl− pump, and photochromic sensor, respectively. Dashed rectangles indicate the positions of NDQ and NTQ motifs and residues unique for XeR. (b) Phylogenetic tree of selected microbial rhodopsins. The rhodopsins included in the phylogenetic tree: bacteriorhodopsin from Halobacterium salinarum (BR), archaerhodopsin-1, -2 and -3 from Halorubrum sodomense (AR1, AR2 and AR3), bacteriorhodopsin and middle rhodopsin from Haloquadratum walsbyi (HwBR, MR), halorhodopsin from H. salinarum, Salinibacter ruber and Natronomonas pharaonis (HsHR, SrHR, NpHR), PoXeR, xenorhodopsin from Haloplasma contractile (HcXeR), ASR, sensory rhodopsin II from N. pharaonis and H. salinarum (NpSRII and HsSRII), sensory rhodopsin I from S. ruber, Haloarcula vallismortis and H. salinarum (SrSRI, HvSRI and HsSRI), proteorhodopsin from Krokinobacter eikastus, Gillisia limnaea, Nonlabens dokdonensis and Vibrio sp. AND4 (KR1, GlPR, NdPR, VsPR), blue-absorbing proteorhodopsin from uncultured bacterium (BPR), green-absorbing proteorhodopsin from uncultured marine gamma proteobacterium (GPR), rhodopsin from Exiguobacterium sibiricum (ESR), thermophilic rhodopsin from Thermus thermophilus (TR), xanthorhodopsin from S. ruber (XR), rhodopsin from Gloeobacter violaceus PCC 7421 (GR), putative Cl− pumping rhodopsin from Citromicrobium sp. JLT1363, Citromicrobium bathyomarinum and Sphingopyxis baekryungensis (CsClR, CbClR, SpClR), Cl- pump from Fulvimarina pelagi and Nonlabens marinus S1-08 (FR and NmClR), PoClR, PoNaR, two putative NaRs from Truepera radiovictrix (TrNaR1 and TrNaR2), putative NaR from Indibacter alkaliphilus (IaNaR), NaR from N. dokdonensis, G. limnaea and K. eikastus (NdNaR, GlNaR and KR2).