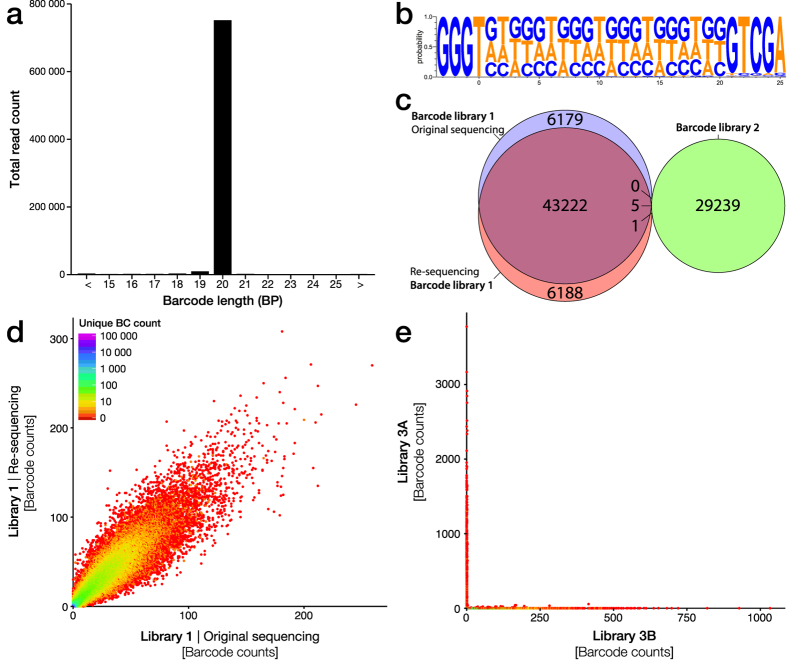
Figure 4. Characterization of molecular barcode diversity.
(a) Length analysis of unfiltered sequencing data from the Ion Torrent platform from library 1 after trimming of the flanking sequences of the degenerate barcodes. This revealed that the vast majority of barcodes are synthesized to the specification of 20 bp. (b) As homopolymer sequences commonly result in sequencing errors in the form of insertion/deletion errors, a cycling nucleotide exclusion paradigm (IUPAC ambiguity code VHDBx5) was used during degenerate primer synthesis to avoid homopolymers longer than three nucleotides. This resulted in close to uniform distribution of the three nucleotides per position with a slight bias for the Guanine, visualized by Weblogo3.3 based on the first 40 000 unique barcodes in library 1. (c) Reproducibility in barcode identification was assessed through comparison of the original Ion Torrent sequencing of library 1 with the re-sequencing conducted using the MiSeq platform and compared those unique barcodes to those found in the library 2 sequenced using the Ion Torrent platform. (d) Analysis of the sequence/re-sequence correlation of the read count per unique barcode for library 1. (e) Using two larger libraries (library 3A and 3B using identical cloning but in two separate reactions) we found near complete orthogonality meaning that each unique barcode was only found in one or the other library. These libraries are estimated to contain around 45 000 and 43 500 unique clones each.