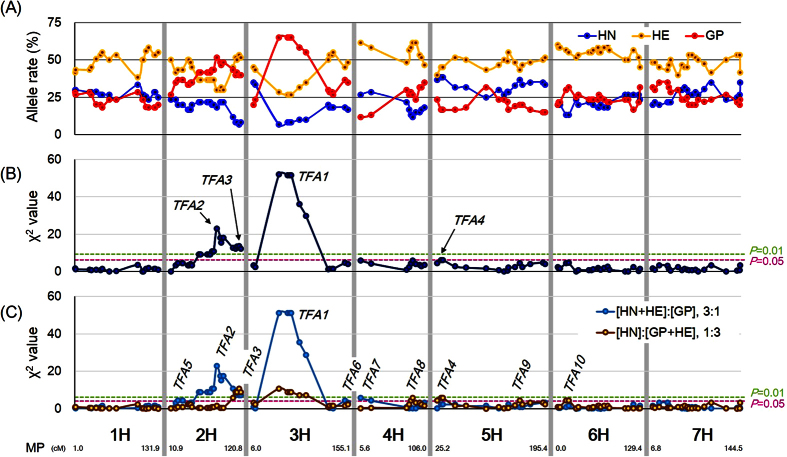
Figure 2. Allele and segregation analysis of SNP markers in transgenic HN × GP plants.
Allele segregation (A) and Chi-square (χ2) values (B,C) were calculated by genotyping 124 SNP markers in transgenic HN × GP plants. SNP markers are shown from short arm (left) to long arm (right) in each chromosome according to the order and genetic distance of markers in genome information of barley1,19. The number (cM) on bottom shows the end marker position (MP) in each chromosome. (A) Blue, yellow, and red line plots show the percentage of ‘Haruna Nijo’ (HN), heterozygous (HE) and Golden Promise (GP) alleles for each SNP marker. (B) Dark blue line plots show χ2 values calculated using 1:2:1 as the expected segregation ratio. Lime green and magenta dotted lines indicate statistical significance at the 1% and 5% levels, respectively (df = 2). (C) Light blue and brown line plots show χ2 values calculated using 3:1 as the expected segregation ratios for [HN + HE]:[GP] and [GP + HE]:[HN], respectively. Lime green and magenta dotted lines indicate statistical significance at the 1% and 5% levels, respectively (df = 1). TFAs (TFA1-10) indicate the loci showing segregation distortion.