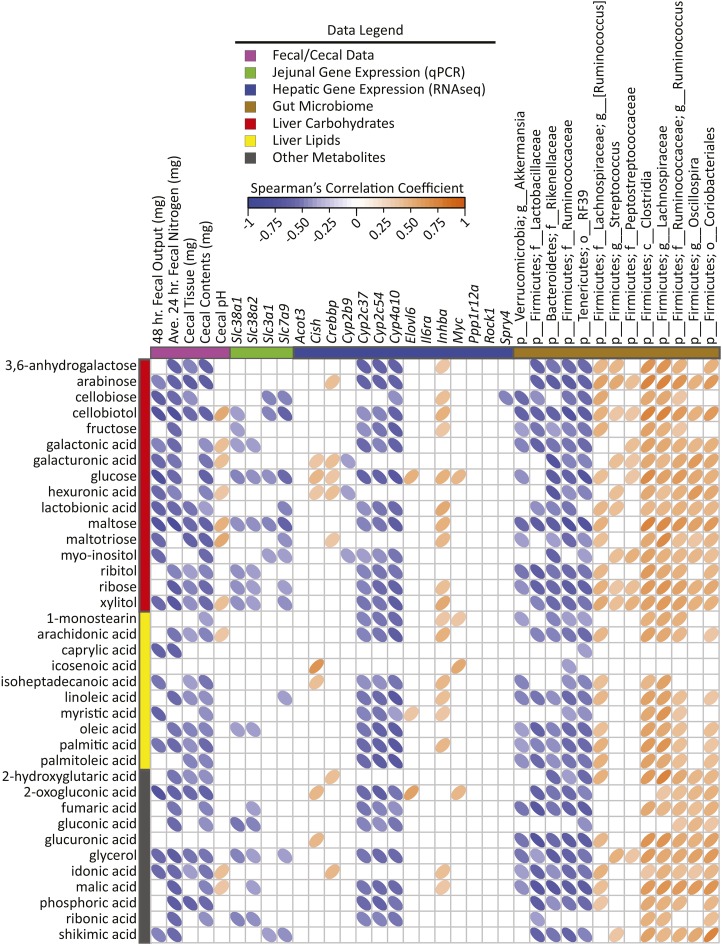
FIGURE 4.
Spearman’s correlation matrix of fecal/cecal data, hepatic gene expression, and cecal bacteria abundances compared with liver carbohydrates, lipids, and miscellaneous metabolites selected in PLS-DA models of male mice fed a 45%-fat diet with or without HAMRS2 supplementation for 10 wk. Bacteria are listed to the lowest level of classification (i.e., if the last taxon assignment is f_, “family” is the lowest level of classification). Metabolites were selected on the basis of having a mean bootstrapped VIP ≥1. The direction of ellipses represents positive or negative correlations and the width of the ellipse represents the strength of correlation (narrow ellipse = stronger correlation). Acot3, acyl-CoA thioesterase 3; Ave., average; c_, class; Cish, cytokine inducible SH2-containing protein; Crebbp, CREB binding protein; Cyp, cytochrome P450; Elovl6, ELOVL family member 6, elongation of long-chain FAs (yeast); f_, family; g_, genus; HAMRS2, high-amylose-maize resistant starch type 2; Il6ra, IL-6 receptor α Inhba, inhibin β-A; Myc, myelocytomatosis oncogene; o_, order; p_, phylum; PLS-DA, partial least-squares-discriminant analysis; Ppp1r12a, protein phosphatase 1, regulatory (inhibitor) subunit 12A; Rock1, rho-associated coiled-coil containing protein kinase 1; Slc, solute carrier; Spry4, sprouty homolog 4 (Drosophila); VIP, variable importance in projection.